ZFLNCG10445
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800043 | sphere stage | normal | 9.21 |
| SRR800046 | sphere stage | 5azaCyD treatment | 8.91 |
| SRR800049 | sphere stage | control treatment | 6.65 |
| SRR800037 | egg | normal | 4.49 |
| SRR372787 | 2-4 cell | normal | 0.98 |
| SRR527835 | tail | normal | 0.68 |
| SRR1049814 | 256 cell | normal | 0.64 |
| SRR1621206 | embryo | normal | 0.60 |
| ERR023143 | swim bladder | normal | 0.47 |
| SRR372791 | dome | normal | 0.42 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| plekhm2 | 0.54 |
| rpusd1 | 0.52 |
| LOC100535847 | 0.51 |
| heatr5a | 0.51 |
| foxh1 | 0.51 |
| zgc:92781 | 0.50 |
| dnal4a | 0.50 |
| si:ch211-132b12.8 | 0.50 |
| zdhhc18b | 0.50 |
Gene Ontology
Download
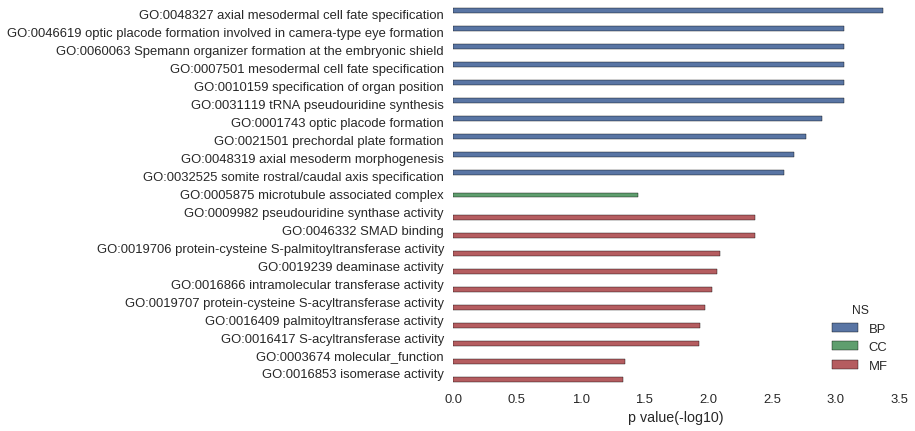
| GO | P value |
|---|---|
| GO:0048327 | 4.26e-04 |
| GO:0046619 | 8.53e-04 |
| GO:0060063 | 8.53e-04 |
| GO:0007501 | 8.53e-04 |
| GO:0010159 | 8.53e-04 |
| GO:0031119 | 8.53e-04 |
| GO:0001743 | 1.28e-03 |
| GO:0021501 | 1.70e-03 |
| GO:0048319 | 2.13e-03 |
| GO:0032525 | 2.56e-03 |
| GO:0005875 | 3.57e-02 |
| GO:0009982 | 4.26e-03 |
| GO:0046332 | 4.26e-03 |
| GO:0019706 | 8.08e-03 |
| GO:0019239 | 8.50e-03 |
| GO:0016866 | 9.35e-03 |
| GO:0019707 | 1.06e-02 |
| GO:0016409 | 1.15e-02 |
| GO:0016417 | 1.19e-02 |
| GO:0003674 | 4.49e-02 |
| GO:0016853 | 4.64e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10445
TCTTGCGTTGTGAGCTATGTTCTGATTTAATTTTTAACACGCTAATTGAAAGTTTTATTTAAGTTGGGTT
GTCTTGCCCAGGTTGTCCATCATGGTGGATACTGGTGGAAACCACATTATACTTCCACTTTTAAAGATTA
TTTTTATTTTTATTACGATAATCACTGAAAAATCTCTTAGGTTGTCTTATGATTCTTCTTGTATTCAATA
AACTTTTGTCATATTCTTAACTGACAGGTCACTTTTCACAAGAGGATCTACTACAATAAGGAGGACAACT
GGAGTCTAGCACTCACCATCTCCAAAGTGCTATATGCTCTCTGTTTGCAGACTGTAACCCTGGTAAGTCA
CACGTCCATAAACACTTCAAGCTTATCGGTACTTGTTAGTTGGTCATGTGTTGATGCTCTGGTGTCTGTC
AGTATGTTGGAGCCTAAGCTGTTTGACCTAGACTAAAACTGAGGCAGTTACAGGTGTGGGCAATTTTGCA
GCAGCTTGATGTTCATGTATAAAACTGCATACACTATTTAGTTTCTTGTGTAAGTGTTAATGATACAGTT
CTTATAGACCGAACGCCACTGTTCCAAAATTTTAAAGCAAGATTTTCAAGTCCAAATCCCATAAAATGGG
CTTTATCGATTTAATCTTGAAAAGGAACCCTCTTGAAAACAATTGAGAGAAGAATGGCATCTATGATTTT
GTCTTTGTCCAGCCAGTATCCAGCCTAGATGATCGATCTGCACGGAGGACAATCACAACAGTGCTATACA
ATCAGAGGACATTCATAAAAGAAGAACCTATCAGTGATGAGACCAATCAGATCTTCATGGCCAACTTAAA
CTGTGTCCTGCTACTTCACCAAGAAGGACAGAAAACTGTGTAAATCTAGTCCCAGTCCAGTTTCCTGGTT
TCCTTCAGTTTTTCACCTCCTCGCAATAAGTTTTAACCTAAGACTACTGAATGTGTTGCAATCTGCTTTG
ACAAGTGGCTTTATGTGAAGCACTATAGAAAAGCATGTGATTGCATCCTTGG
TCTTGCGTTGTGAGCTATGTTCTGATTTAATTTTTAACACGCTAATTGAAAGTTTTATTTAAGTTGGGTT
GTCTTGCCCAGGTTGTCCATCATGGTGGATACTGGTGGAAACCACATTATACTTCCACTTTTAAAGATTA
TTTTTATTTTTATTACGATAATCACTGAAAAATCTCTTAGGTTGTCTTATGATTCTTCTTGTATTCAATA
AACTTTTGTCATATTCTTAACTGACAGGTCACTTTTCACAAGAGGATCTACTACAATAAGGAGGACAACT
GGAGTCTAGCACTCACCATCTCCAAAGTGCTATATGCTCTCTGTTTGCAGACTGTAACCCTGGTAAGTCA
CACGTCCATAAACACTTCAAGCTTATCGGTACTTGTTAGTTGGTCATGTGTTGATGCTCTGGTGTCTGTC
AGTATGTTGGAGCCTAAGCTGTTTGACCTAGACTAAAACTGAGGCAGTTACAGGTGTGGGCAATTTTGCA
GCAGCTTGATGTTCATGTATAAAACTGCATACACTATTTAGTTTCTTGTGTAAGTGTTAATGATACAGTT
CTTATAGACCGAACGCCACTGTTCCAAAATTTTAAAGCAAGATTTTCAAGTCCAAATCCCATAAAATGGG
CTTTATCGATTTAATCTTGAAAAGGAACCCTCTTGAAAACAATTGAGAGAAGAATGGCATCTATGATTTT
GTCTTTGTCCAGCCAGTATCCAGCCTAGATGATCGATCTGCACGGAGGACAATCACAACAGTGCTATACA
ATCAGAGGACATTCATAAAAGAAGAACCTATCAGTGATGAGACCAATCAGATCTTCATGGCCAACTTAAA
CTGTGTCCTGCTACTTCACCAAGAAGGACAGAAAACTGTGTAAATCTAGTCCCAGTCCAGTTTCCTGGTT
TCCTTCAGTTTTTCACCTCCTCGCAATAAGTTTTAACCTAAGACTACTGAATGTGTTGCAATCTGCTTTG
ACAAGTGGCTTTATGTGAAGCACTATAGAAAAGCATGTGATTGCATCCTTGG

