ZFLNCG10523
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647683 | spleen | SVCV treatment | 9.27 |
| SRR1647680 | head kidney | SVCV treatment | 7.43 |
| SRR748488 | sperm | normal | 6.61 |
| SRR1647679 | head kidney | normal | 6.43 |
| SRR1647682 | spleen | normal | 4.92 |
| SRR1647684 | spleen | SVCV treatment | 3.92 |
| SRR1562532 | spleen | normal | 3.52 |
| SRR1647681 | head kidney | SVCV treatment | 3.47 |
| ERR023144 | brain | normal | 2.75 |
| ERR023146 | head kidney | normal | 2.55 |
Express in tissues
Correlated coding gene
Download
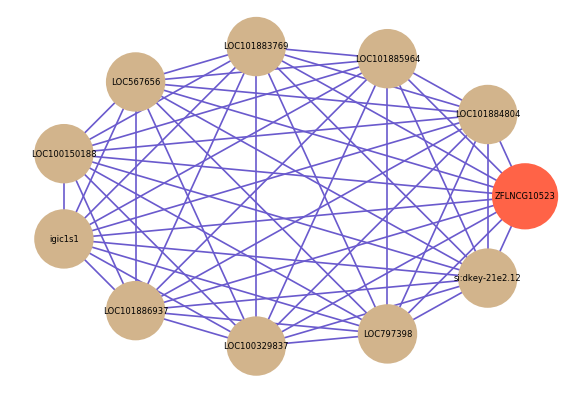
| gene | correlation coefficent |
|---|---|
| LOC101884804 | 0.66 |
| LOC101885964 | 0.66 |
| LOC101883769 | 0.65 |
| LOC567656 | 0.65 |
| LOC100150188 | 0.64 |
| igic1s1 | 0.62 |
| LOC101886937 | 0.62 |
| LOC100329837 | 0.60 |
| LOC797398 | 0.60 |
| si:dkey-21e2.12 | 0.60 |
Gene Ontology
Download
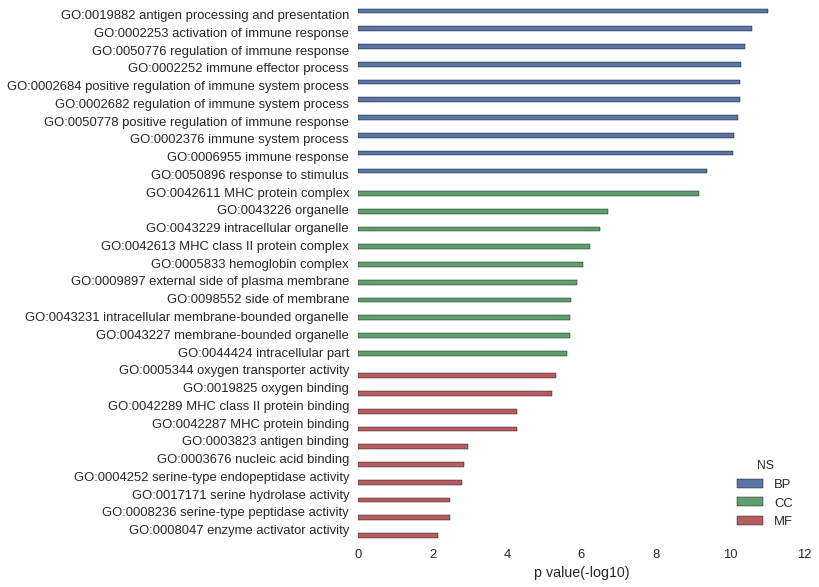
| GO | P value |
|---|---|
| GO:0019882 | 9.48e-12 |
| GO:0002253 | 2.65e-11 |
| GO:0050776 | 3.99e-11 |
| GO:0002252 | 4.99e-11 |
| GO:0002684 | 5.47e-11 |
| GO:0002682 | 5.59e-11 |
| GO:0050778 | 6.11e-11 |
| GO:0002376 | 7.66e-11 |
| GO:0006955 | 8.56e-11 |
| GO:0050896 | 4.23e-10 |
| GO:0042611 | 6.68e-10 |
| GO:0043226 | 1.98e-07 |
| GO:0043229 | 3.21e-07 |
| GO:0042613 | 5.71e-07 |
| GO:0005833 | 8.93e-07 |
| GO:0009897 | 1.33e-06 |
| GO:0098552 | 1.91e-06 |
| GO:0043231 | 2.07e-06 |
| GO:0043227 | 2.08e-06 |
| GO:0044424 | 2.37e-06 |
| GO:0005344 | 4.79e-06 |
| GO:0019825 | 6.22e-06 |
| GO:0042289 | 5.41e-05 |
| GO:0042287 | 5.41e-05 |
| GO:0003823 | 1.11e-03 |
| GO:0003676 | 1.47e-03 |
| GO:0004252 | 1.61e-03 |
| GO:0017171 | 3.35e-03 |
| GO:0008236 | 3.35e-03 |
| GO:0008047 | 7.18e-03 |
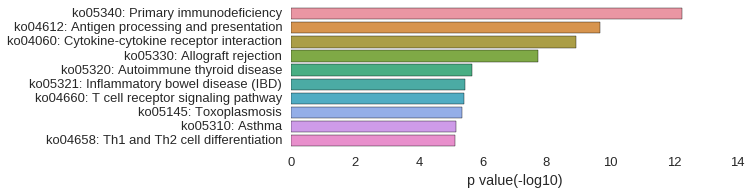
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10523
GAACACCCAGTTATCACTGATCTTATGGACACTCTGACAGTAATAATCTCCTGCATCTTCATGCTGAGCT
TCACTGATGCTCAAAGTGAAGTCAAGACCGTTTCCCTGTCCACTGCCACTGAATCGAGATGGAATTCCTG
AGTTTTTTTCGCTTGTGTAATAAATCAAGAGTCTGGGAGCCTCTTCAGGCTTTTGGCTGTACCAGTGTAT
ATAATAACGGCCTTTGGAGGTGGTGCCTTGATCATTTTTAAAAACCACAGTATTGACTTTACAGTTTATA
GAGACTGTTTGACCAGGCTGGACTGATTTAACCTCATACTGAGTCACAACAATTTGTCCAAAGCAACCTG
TGTAGAACAAGAAATAACAGCACTGAATAATTACATTAATACTGAAATATATTTTAAAGTTATTTGTAAT
AATGTTTTTATTTTACCTTGTAGGCTGTAGAGCATCCAGATGATAAAGGACATAAAAGTCAT
GAACACCCAGTTATCACTGATCTTATGGACACTCTGACAGTAATAATCTCCTGCATCTTCATGCTGAGCT
TCACTGATGCTCAAAGTGAAGTCAAGACCGTTTCCCTGTCCACTGCCACTGAATCGAGATGGAATTCCTG
AGTTTTTTTCGCTTGTGTAATAAATCAAGAGTCTGGGAGCCTCTTCAGGCTTTTGGCTGTACCAGTGTAT
ATAATAACGGCCTTTGGAGGTGGTGCCTTGATCATTTTTAAAAACCACAGTATTGACTTTACAGTTTATA
GAGACTGTTTGACCAGGCTGGACTGATTTAACCTCATACTGAGTCACAACAATTTGTCCAAAGCAACCTG
TGTAGAACAAGAAATAACAGCACTGAATAATTACATTAATACTGAAATATATTTTAAAGTTATTTGTAAT
AATGTTTTTATTTTACCTTGTAGGCTGTAGAGCATCCAGATGATAAAGGACATAAAAGTCAT