ZFLNCG10718
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941753 | posterior pectoral fin | normal | 65.84 |
| SRR941749 | anterior pectoral fin | normal | 58.24 |
| SRR891495 | heart | normal | 50.64 |
| SRR516128 | skin | male and 5 month | 28.97 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 25.21 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 19.63 |
| SRR516129 | skin | male and 5 month | 16.75 |
| SRR516131 | skin | male and 5 month | 14.44 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 14.05 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 13.84 |
Express in tissues
Correlated coding gene
Download
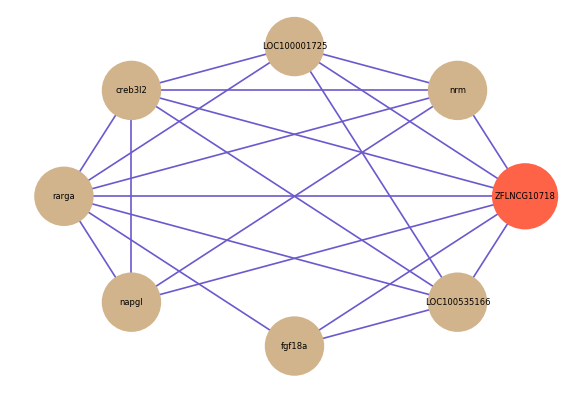
| gene | correlation coefficent |
|---|---|
| nrm | 0.55 |
| fgf18a | 0.53 |
| LOC100001725 | 0.52 |
| creb3l2 | 0.51 |
| rarga | 0.51 |
| LOC100535166 | 0.50 |
| napgl | 0.50 |
Gene Ontology
Download
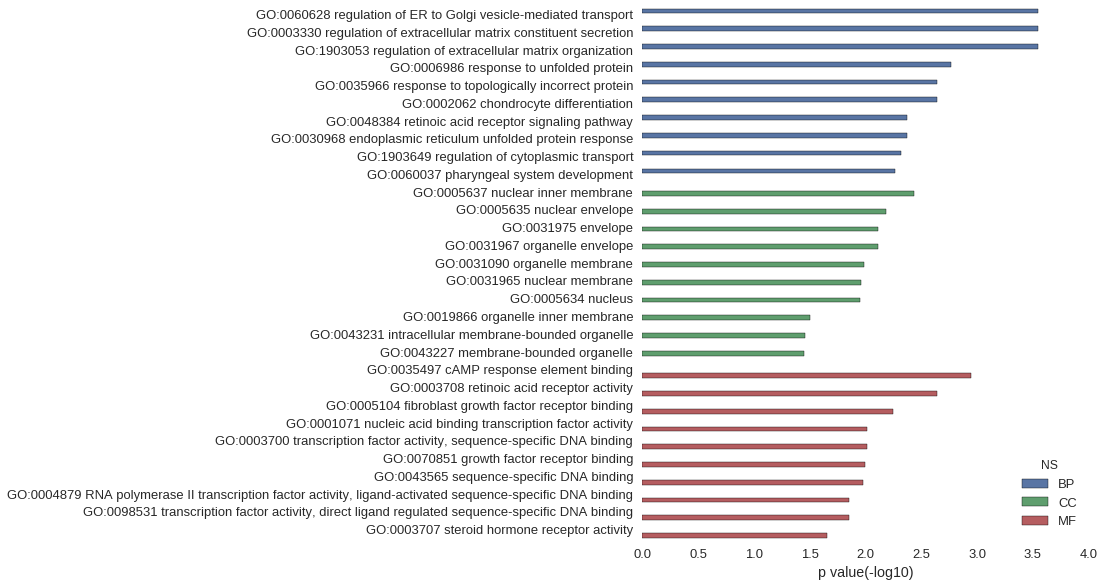
| GO | P value |
|---|---|
| GO:0060628 | 2.84e-04 |
| GO:0003330 | 2.84e-04 |
| GO:1903053 | 2.84e-04 |
| GO:0006986 | 1.70e-03 |
| GO:0035966 | 2.27e-03 |
| GO:0002062 | 2.27e-03 |
| GO:0048384 | 4.26e-03 |
| GO:0030968 | 4.26e-03 |
| GO:1903649 | 4.82e-03 |
| GO:0060037 | 5.39e-03 |
| GO:0005637 | 3.69e-03 |
| GO:0005635 | 6.52e-03 |
| GO:0031975 | 7.65e-03 |
| GO:0031967 | 7.65e-03 |
| GO:0031090 | 1.03e-02 |
| GO:0031965 | 1.10e-02 |
| GO:0005634 | 1.11e-02 |
| GO:0019866 | 3.12e-02 |
| GO:0043231 | 3.51e-02 |
| GO:0043227 | 3.57e-02 |
| GO:0035497 | 1.14e-03 |
| GO:0003708 | 2.27e-03 |
| GO:0005104 | 5.67e-03 |
| GO:0001071 | 9.72e-03 |
| GO:0003700 | 9.72e-03 |
| GO:0070851 | 1.02e-02 |
| GO:0043565 | 1.06e-02 |
| GO:0004879 | 1.41e-02 |
| GO:0098531 | 1.41e-02 |
| GO:0003707 | 2.20e-02 |
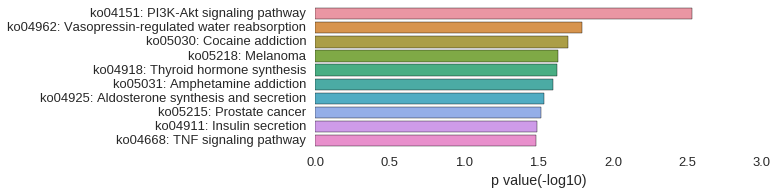
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10718
CTGGATTCCACGAGCTCCTCAGCAGTTTCCCAGCAGACAGTCCTTTCGTCAGAGAGACGCCCGGCAGACC
TGCCAACTGCACCCAGCGCTTCTGGCTGCCACCATCCTCCCCGGTGTGCTGGGATGACATCGCGGGACCA
GAGGAGTTTGAGAAGTCCCGTATGCTGGTGCTGCAGAACCGGGCCGCTCTGCAGGCCGTGTCCACATCCA
GCGGCCTGGAGGATGGAGGAGTGTCTTACGATCAGCAGGCCAGAGAGGAGGTGCAGGGCGTGCGGGAAGA
TCACCTTGCAGTCATCCAGACCACAGACACCATGCAGAAAGTCTTCCTGGATCTGGAGGAGAAGAGGAAG
GAGGGGAAAGAGCATTATACCTTCAACAGGTGAATGAGAACCTTTATTTCTGGGGGAAGATATGTGACAC
ATGAGCAAATCCAGCATCATGGATGCAGCTAAAACCCAAACATGAATACACAGAGAAAGTATAGGTTAAC
ATTATATTGAAATACCTGTTTAGCCG
CTGGATTCCACGAGCTCCTCAGCAGTTTCCCAGCAGACAGTCCTTTCGTCAGAGAGACGCCCGGCAGACC
TGCCAACTGCACCCAGCGCTTCTGGCTGCCACCATCCTCCCCGGTGTGCTGGGATGACATCGCGGGACCA
GAGGAGTTTGAGAAGTCCCGTATGCTGGTGCTGCAGAACCGGGCCGCTCTGCAGGCCGTGTCCACATCCA
GCGGCCTGGAGGATGGAGGAGTGTCTTACGATCAGCAGGCCAGAGAGGAGGTGCAGGGCGTGCGGGAAGA
TCACCTTGCAGTCATCCAGACCACAGACACCATGCAGAAAGTCTTCCTGGATCTGGAGGAGAAGAGGAAG
GAGGGGAAAGAGCATTATACCTTCAACAGGTGAATGAGAACCTTTATTTCTGGGGGAAGATATGTGACAC
ATGAGCAAATCCAGCATCATGGATGCAGCTAAAACCCAAACATGAATACACAGAGAAAGTATAGGTTAAC
ATTATATTGAAATACCTGTTTAGCCG