ZFLNCG10762
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 7.05 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 6.71 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 5.44 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 5.23 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 4.93 |
| SRR891511 | brain | normal | 3.99 |
| SRR516131 | skin | male and 5 month | 3.90 |
| SRR516122 | skin | male and 3.5 year | 3.86 |
| SRR516125 | skin | male and 3.5 year | 3.82 |
| SRR516135 | skin | male and 5 month | 3.67 |
Express in tissues
Correlated coding gene
Download
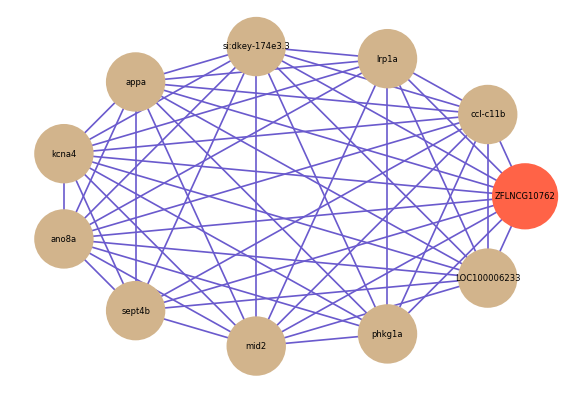
| gene | correlation coefficent |
|---|---|
| ccl-c11b | 0.62 |
| lrp1a | 0.62 |
| si:dkey-174e3.3 | 0.62 |
| appa | 0.60 |
| kcna4 | 0.60 |
| ano8a | 0.59 |
| sept4b | 0.59 |
| mid2 | 0.59 |
| phkg1a | 0.59 |
| LOC100006233 | 0.58 |
Gene Ontology
Download
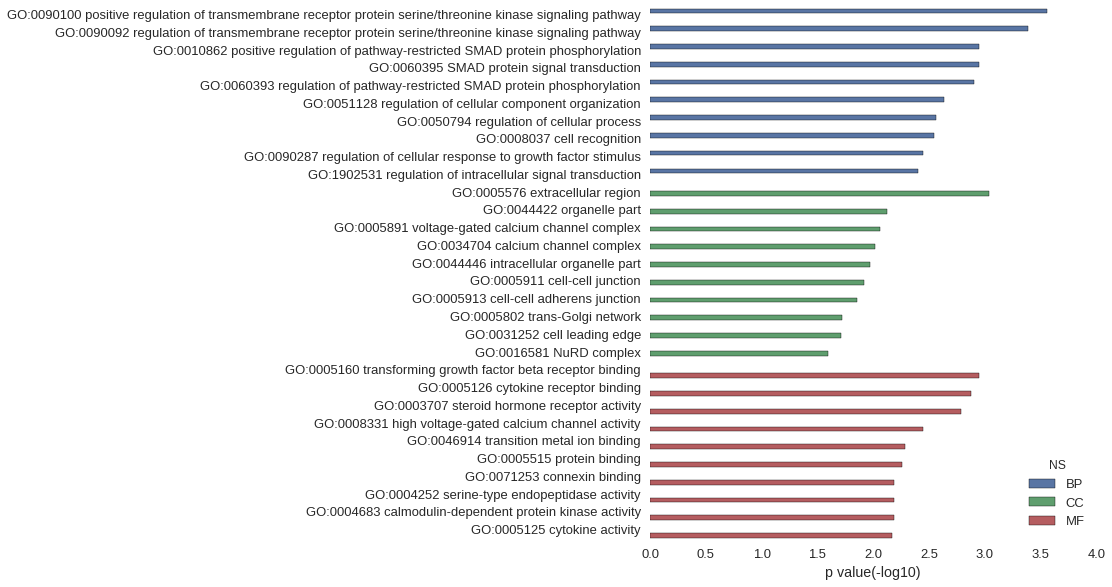
| GO | P value |
|---|---|
| GO:0090100 | 2.78e-04 |
| GO:0090092 | 4.08e-04 |
| GO:0010862 | 1.13e-03 |
| GO:0060395 | 1.13e-03 |
| GO:0060393 | 1.24e-03 |
| GO:0051128 | 2.34e-03 |
| GO:0050794 | 2.75e-03 |
| GO:0008037 | 2.86e-03 |
| GO:0090287 | 3.59e-03 |
| GO:1902531 | 4.00e-03 |
| GO:0005576 | 9.26e-04 |
| GO:0044422 | 7.55e-03 |
| GO:0005891 | 8.78e-03 |
| GO:0034704 | 9.58e-03 |
| GO:0044446 | 1.06e-02 |
| GO:0005911 | 1.20e-02 |
| GO:0005913 | 1.40e-02 |
| GO:0005802 | 1.92e-02 |
| GO:0031252 | 1.93e-02 |
| GO:0016581 | 2.56e-02 |
| GO:0005160 | 1.13e-03 |
| GO:0005126 | 1.33e-03 |
| GO:0003707 | 1.62e-03 |
| GO:0008331 | 3.58e-03 |
| GO:0046914 | 5.20e-03 |
| GO:0005515 | 5.50e-03 |
| GO:0071253 | 6.47e-03 |
| GO:0004252 | 6.56e-03 |
| GO:0004683 | 6.58e-03 |
| GO:0005125 | 6.86e-03 |
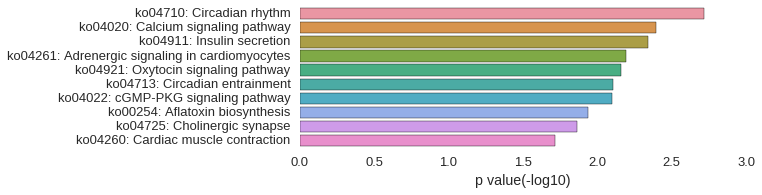
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10762
CTGGAGTCATTGATGGCTAATCTGAGCATCTCGCCCTCATTCACACACACTGATTCACAGTTCTCTCAGA
GAGAGAGAGTATGGTGAGAGTTTCTGTTTATGTGTTTCTGTTTATAAGTGTGTCTGTGATGGAGAGAGAG
AGAGAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCT
GCAGCTCCTGCGTTTTTCCTCAGTCACCATAGAGAAATGCAAAGTTGAAGTTGAAAACAAGGAAAGGCCT
TGGTCTCCATGGCAACAGCAGTGCCTTAGTAAACAAAGAGAAAGATGAGACGGACGTGTGTGTGTGTGTG
TGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGACTCTAAGGGACCGCAGGTATT
TTCTCAGTCCAGTTCCATGATGCATTTCAGCGTGATGTCATCATGTGATCATGTGTCCTCTTCGTCTGAC
GTCTGTCTAAAGAACAAACCCAGCAAATATATTATCCTCAGATAATGAGAAAACAAGAAGCAGTAAAGCG
CGGTGTGTTTCCAGCGCTGAGAAACAAACACACCTCCGCCTTCACTCCGCTGACTCCTG
CTGGAGTCATTGATGGCTAATCTGAGCATCTCGCCCTCATTCACACACACTGATTCACAGTTCTCTCAGA
GAGAGAGAGTATGGTGAGAGTTTCTGTTTATGTGTTTCTGTTTATAAGTGTGTCTGTGATGGAGAGAGAG
AGAGAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCT
GCAGCTCCTGCGTTTTTCCTCAGTCACCATAGAGAAATGCAAAGTTGAAGTTGAAAACAAGGAAAGGCCT
TGGTCTCCATGGCAACAGCAGTGCCTTAGTAAACAAAGAGAAAGATGAGACGGACGTGTGTGTGTGTGTG
TGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGACTCTAAGGGACCGCAGGTATT
TTCTCAGTCCAGTTCCATGATGCATTTCAGCGTGATGTCATCATGTGATCATGTGTCCTCTTCGTCTGAC
GTCTGTCTAAAGAACAAACCCAGCAAATATATTATCCTCAGATAATGAGAAAACAAGAAGCAGTAAAGCG
CGGTGTGTTTCCAGCGCTGAGAAACAAACACACCTCCGCCTTCACTCCGCTGACTCCTG