ZFLNCG10797
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 19.83 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 5.97 |
| SRR891504 | liver | normal | 5.28 |
| SRR658541 | 6 somite | normal | 2.56 |
| SRR658539 | bud | Gata5/6 morphant | 2.47 |
| SRR658543 | 6 somite | Gata5 morphan | 2.28 |
| SRR594769 | blood | normal | 2.24 |
| SRR658545 | 6 somite | Gata6 morphant | 2.19 |
| SRR658533 | bud | normal | 2.02 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 1.99 |
Express in tissues
Correlated coding gene
Download
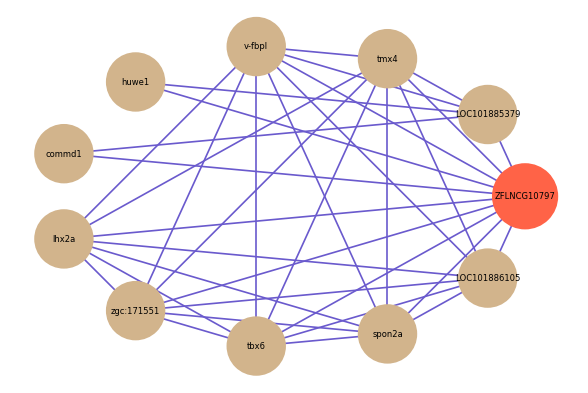
| gene | correlation coefficent |
|---|---|
| LOC101885379 | 0.56 |
| lhx2a | 0.53 |
| zgc:171551 | 0.53 |
| tbx6 | 0.52 |
| tmx4 | 0.52 |
| v-fbpl | 0.52 |
| spon2a | 0.51 |
| LOC101886105 | 0.51 |
| huwe1 | 0.51 |
| commd1 | 0.50 |
Gene Ontology
Download
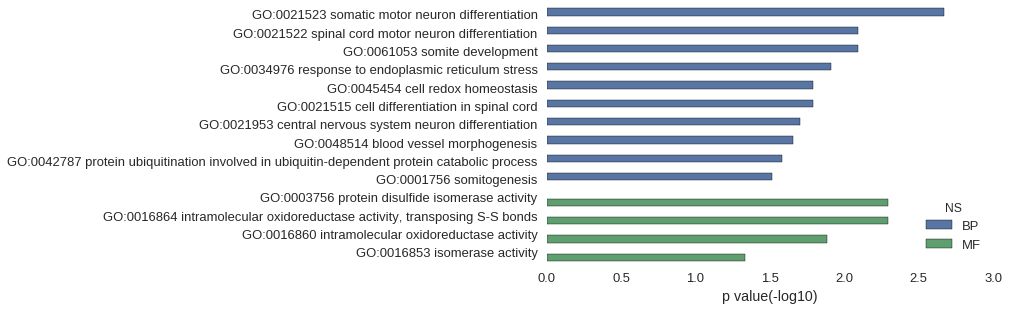
| GO | P value |
|---|---|
| GO:0021523 | 2.13e-03 |
| GO:0021522 | 8.08e-03 |
| GO:0061053 | 8.08e-03 |
| GO:0034976 | 1.23e-02 |
| GO:0045454 | 1.61e-02 |
| GO:0021515 | 1.61e-02 |
| GO:0021953 | 1.99e-02 |
| GO:0048514 | 2.20e-02 |
| GO:0042787 | 2.62e-02 |
| GO:0001756 | 3.07e-02 |
| GO:0003756 | 5.11e-03 |
| GO:0016864 | 5.11e-03 |
| GO:0016860 | 1.31e-02 |
| GO:0016853 | 4.64e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10797
CGCCTGGGAAAAGGCTGAGATCAGCAAGAAGTGGGAGGAGAGCAGCTGGGCCAAGAAGATCGAGGCCAGG
AAGAAGGTAAACACACACACTTACATGAGCAGATTCACACACACACACACACACCTACATGAGCAGATTC
ACACACACACACACACCTACATGAGCAGATTCACACACACACACACACACACACACCTACATGAGCAGAT
TCTCACACACACACACACACACCTACATGAGCAGATTCACACACACACACACACACACCTACATGAGCAG
ATTCTCACACAAACCTATGCAGATTTTCTCTCACACACACACACATGCAGATTCACACGCACAGGAAATG
GAACATGTAAAGCAGCTGATGGTGTTTTTCAGAGTGTTTTAGCTGCAGTATTGTGACGACATCCTATATG
TAAAACACACACTTTTACTGTAAATGCCCCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGC
GTGTGTGTTTAATGATGCGATTTTTCTCTGCATTATTTGAGCTCTTTATGGAGTGAGGAACAGTATATGT
GACCTGGAACACACACACACACACACACACACACCTTATTTTGGCTGTATTTGGCAGCAGACACACATTA
TTCCATGTGTATATAAAAGTGTTTTGTTTTCGGCCAGAAGCAGCAGGTTATGATATTTTGTAGGTATTTT
AACATTAAACTCTCCTTTTGAAGAGTGTTTTGTCAGTGTTTAGACATTTCATATTTAGATATAATATTCC
GCTGGTTCTGTACGCCTCAGATTGCAGATCTTCAGAGTTTTCTCTCCGCTAAACATTGTTTAATCCTCAC
ATTAATGAAAGGATGAGTGATTGAGCTGGTGCATGAAGTGTAATCTCTGTAGACTGTGTGTGACTGAGAC
GTTCTGCTGAACAGAGAGTGTCGGTCCAGTGTTCAGAGCTGAAGTCAATGATACAGCCTGACCTGTTCAC
TGCTATAATATTGTGAATGTTTCTCTGTATAATGCATAGTGTGTTGCTCCTCATGTTTTCACAATGTGAA
GAGCGTTCGCTGTGATCACACCCTAACTCTCCATTTCAAACACTTATCTTTGTCCTGGCGCACGTCTGCT
CTCTTCATCTACATTGATTACAATATCAGACAGTATTGAAAGTAATAAAATAATAATAACTTTATTAACT
TATATTAGTGGT
CGCCTGGGAAAAGGCTGAGATCAGCAAGAAGTGGGAGGAGAGCAGCTGGGCCAAGAAGATCGAGGCCAGG
AAGAAGGTAAACACACACACTTACATGAGCAGATTCACACACACACACACACACCTACATGAGCAGATTC
ACACACACACACACACCTACATGAGCAGATTCACACACACACACACACACACACACCTACATGAGCAGAT
TCTCACACACACACACACACACCTACATGAGCAGATTCACACACACACACACACACACCTACATGAGCAG
ATTCTCACACAAACCTATGCAGATTTTCTCTCACACACACACACATGCAGATTCACACGCACAGGAAATG
GAACATGTAAAGCAGCTGATGGTGTTTTTCAGAGTGTTTTAGCTGCAGTATTGTGACGACATCCTATATG
TAAAACACACACTTTTACTGTAAATGCCCCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGC
GTGTGTGTTTAATGATGCGATTTTTCTCTGCATTATTTGAGCTCTTTATGGAGTGAGGAACAGTATATGT
GACCTGGAACACACACACACACACACACACACACCTTATTTTGGCTGTATTTGGCAGCAGACACACATTA
TTCCATGTGTATATAAAAGTGTTTTGTTTTCGGCCAGAAGCAGCAGGTTATGATATTTTGTAGGTATTTT
AACATTAAACTCTCCTTTTGAAGAGTGTTTTGTCAGTGTTTAGACATTTCATATTTAGATATAATATTCC
GCTGGTTCTGTACGCCTCAGATTGCAGATCTTCAGAGTTTTCTCTCCGCTAAACATTGTTTAATCCTCAC
ATTAATGAAAGGATGAGTGATTGAGCTGGTGCATGAAGTGTAATCTCTGTAGACTGTGTGTGACTGAGAC
GTTCTGCTGAACAGAGAGTGTCGGTCCAGTGTTCAGAGCTGAAGTCAATGATACAGCCTGACCTGTTCAC
TGCTATAATATTGTGAATGTTTCTCTGTATAATGCATAGTGTGTTGCTCCTCATGTTTTCACAATGTGAA
GAGCGTTCGCTGTGATCACACCCTAACTCTCCATTTCAAACACTTATCTTTGTCCTGGCGCACGTCTGCT
CTCTTCATCTACATTGATTACAATATCAGACAGTATTGAAAGTAATAAAATAATAATAACTTTATTAACT
TATATTAGTGGT

