ZFLNCG10803
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR514030 | retina | id2a morpholino | 144.57 |
| SRR514028 | retina | control morpholino | 138.39 |
| SRR1104058 | heart | normal | 29.65 |
| SRR1104059 | heart | kctd10 mut | 12.47 |
| SRR1205166 | 5dpf | transgenic sqET20 and neomycin treated 3h | 11.48 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 10.75 |
| SRR065196 | 3 dpf | normal | 10.52 |
| SRR1205154 | 5dpf | transgenic sqET20 | 9.28 |
| SRR1205172 | 5dpf | transgenic sqET20 and neomycin treated 5h | 8.21 |
| SRR726540 | 5 dpf | normal | 6.55 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| si:dkey-159f12.2 | 0.52 |
Gene Ontology
Download
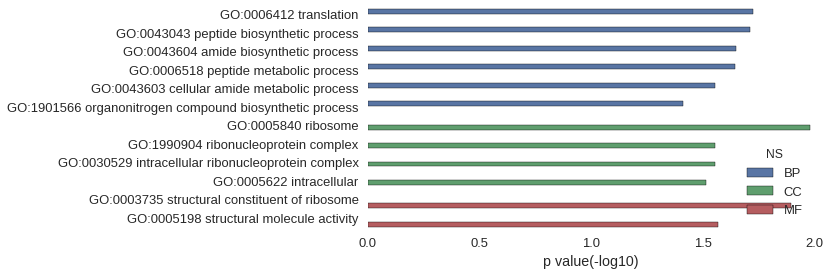
| GO | P value |
|---|---|
| GO:0006412 | 1.88e-02 |
| GO:0043043 | 1.93e-02 |
| GO:0043604 | 2.25e-02 |
| GO:0006518 | 2.27e-02 |
| GO:0043603 | 2.79e-02 |
| GO:1901566 | 3.88e-02 |
| GO:0005840 | 1.04e-02 |
| GO:1990904 | 2.79e-02 |
| GO:0030529 | 2.79e-02 |
| GO:0005622 | 3.06e-02 |
| GO:0003735 | 1.27e-02 |
| GO:0005198 | 2.70e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10803
TTCACTTTTAAAATAGGTGATATAACACATTTAAACACACACATTGTACTCACTTTAAGATTGGAGAGCT
GTCCGAAAGTCTTCATACAGACATTGCATTCATATTTGATCTTCCCGTTCTGCTTTTTCAGTGGATACGG
GAGAGATTTATACCCAGGAATCATGGCTGGAGAGGCTGACGGAGGCGGTGTGCTCTTAGGTGTAGTCAGA
CTCAGATTTATGGCTTCTTCACATTCAGAATAATGCGTCTGGCAAGGAGAAGTCTGAGGGGAAAGTTCTG
GCGTTGCAGGCGCTCCTCTGGGGGGAGTATTGTGCCCCCTTTCGTTTAATCCTCCGTGGAGTGTGTTAGG
ATATGGAAGAGATACAGGTAATAAACCTGACTGTGGTATTGAAGGATAGGACAAACCGTCAGGATTCATG
AAGCCGTATCGGTGGGCTGGTTTTGTGGGAAGCATTGAAGTGAACGGAGGAGAGTATGGAGGAATGAGCA
TGTGCTGGTAATGAGGGTTATACTGCGGTAAGAGCGGATATGGAGACCGTATTGACGGATATGAGGAGAT
CAAACCCTCTCGGTGACCGTACAGGCTGTGTAGATGCAGCGGGACGTCCTGCGGTGGAGACAGGCTCGGG
TAAATGTTTGTTTTTTCATGCATTGGTTCTGACTTGTGGATCTTCTTGCTGAAGTCCATTATTTGCTCGT
CTGGAGTGTCTGGCGGCGTGTCTCTCTCGATGTCGATCTTCTCTTCAGGTTCCTCGTCGTCTTTATTTGT
GTCGCAGGGATGGTTTTGTGGGTTCGAGGGGTGTTTGGGGTAATATGTGTACTGAGGTAGAGGAGGCTTC
TGGTACTCCATATCTGATG
TTCACTTTTAAAATAGGTGATATAACACATTTAAACACACACATTGTACTCACTTTAAGATTGGAGAGCT
GTCCGAAAGTCTTCATACAGACATTGCATTCATATTTGATCTTCCCGTTCTGCTTTTTCAGTGGATACGG
GAGAGATTTATACCCAGGAATCATGGCTGGAGAGGCTGACGGAGGCGGTGTGCTCTTAGGTGTAGTCAGA
CTCAGATTTATGGCTTCTTCACATTCAGAATAATGCGTCTGGCAAGGAGAAGTCTGAGGGGAAAGTTCTG
GCGTTGCAGGCGCTCCTCTGGGGGGAGTATTGTGCCCCCTTTCGTTTAATCCTCCGTGGAGTGTGTTAGG
ATATGGAAGAGATACAGGTAATAAACCTGACTGTGGTATTGAAGGATAGGACAAACCGTCAGGATTCATG
AAGCCGTATCGGTGGGCTGGTTTTGTGGGAAGCATTGAAGTGAACGGAGGAGAGTATGGAGGAATGAGCA
TGTGCTGGTAATGAGGGTTATACTGCGGTAAGAGCGGATATGGAGACCGTATTGACGGATATGAGGAGAT
CAAACCCTCTCGGTGACCGTACAGGCTGTGTAGATGCAGCGGGACGTCCTGCGGTGGAGACAGGCTCGGG
TAAATGTTTGTTTTTTCATGCATTGGTTCTGACTTGTGGATCTTCTTGCTGAAGTCCATTATTTGCTCGT
CTGGAGTGTCTGGCGGCGTGTCTCTCTCGATGTCGATCTTCTCTTCAGGTTCCTCGTCGTCTTTATTTGT
GTCGCAGGGATGGTTTTGTGGGTTCGAGGGGTGTTTGGGGTAATATGTGTACTGAGGTAGAGGAGGCTTC
TGGTACTCCATATCTGATG
