ZFLNCG10888
Basic Information
Chromesome: chr20
Start: 10676544
End: 10677150
Transcript: ZFLNCT16855 ZFLNCT16856
Known as: ENSDARG00000097342 LOC101882890
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800045 | muscle | normal | 254.35 |
| SRR800037 | egg | normal | 136.04 |
| SRR800046 | sphere stage | 5azaCyD treatment | 135.05 |
| SRR800043 | sphere stage | normal | 111.51 |
| SRR800049 | sphere stage | control treatment | 94.34 |
| SRR748488 | sperm | normal | 22.48 |
| SRR658533 | bud | normal | 20.46 |
| SRR658537 | bud | Gata6 morphant | 17.11 |
| SRR658545 | 6 somite | Gata6 morphant | 13.88 |
| SRR749514 | 24 hpf | normal | 13.34 |
Express in tissues
Correlated coding gene
Download
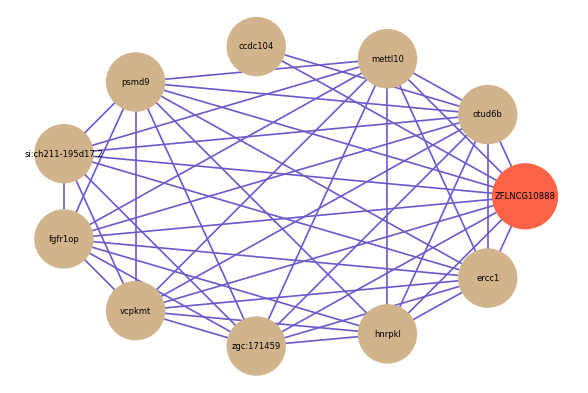
| gene | correlation coefficent |
|---|---|
| otud6b | 0.55 |
| mettl10 | 0.55 |
| ccdc104 | 0.54 |
| psmd9 | 0.54 |
| si:ch211-195d17.2 | 0.54 |
| fgfr1op | 0.54 |
| vcpkmt | 0.52 |
| zgc:171459 | 0.51 |
| hnrpkl | 0.51 |
| ercc1 | 0.50 |
Gene Ontology
Download
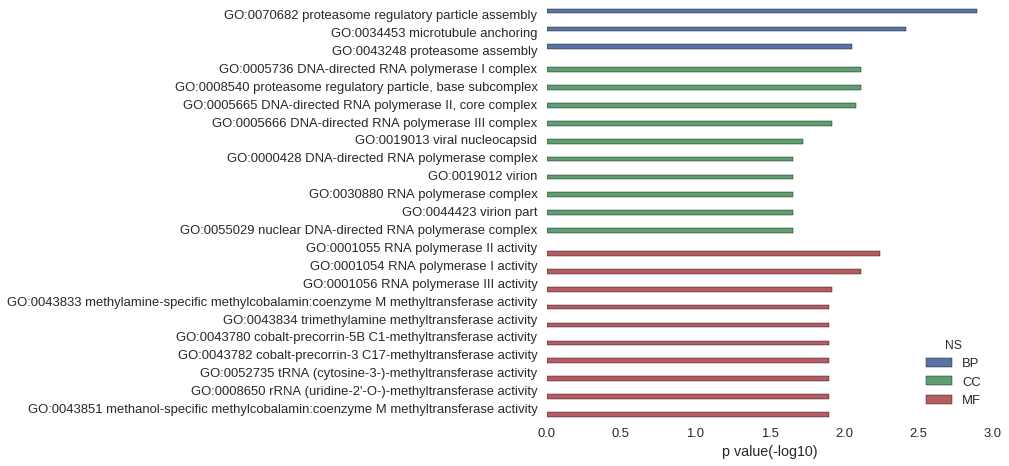
| GO | P value |
|---|---|
| GO:0070682 | 1.28e-03 |
| GO:0034453 | 3.83e-03 |
| GO:0043248 | 8.92e-03 |
| GO:0005736 | 7.65e-03 |
| GO:0008540 | 7.65e-03 |
| GO:0005665 | 8.29e-03 |
| GO:0005666 | 1.21e-02 |
| GO:0019013 | 1.90e-02 |
| GO:0000428 | 2.22e-02 |
| GO:0019012 | 2.22e-02 |
| GO:0030880 | 2.22e-02 |
| GO:0044423 | 2.22e-02 |
| GO:0055029 | 2.22e-02 |
| GO:0001055 | 5.74e-03 |
| GO:0001054 | 7.65e-03 |
| GO:0001056 | 1.21e-02 |
| GO:0043833 | 1.27e-02 |
| GO:0043834 | 1.27e-02 |
| GO:0043780 | 1.27e-02 |
| GO:0043782 | 1.27e-02 |
| GO:0052735 | 1.27e-02 |
| GO:0008650 | 1.27e-02 |
| GO:0043851 | 1.27e-02 |
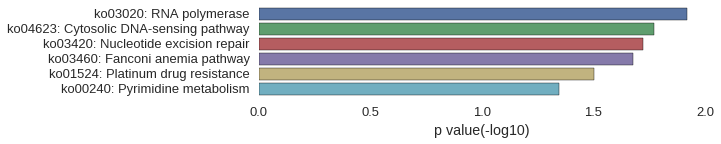
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10888
GATTCGACTCTGAGTGAATGATTCATTCACGAAGCTCTATTTAACTGCACGATGTTGCATGGGCTGAGGA
GACATGTTGTTGAAGGCTGCATAAATGTTACTTGTATGAAGTAAGTAGCTTAAGATAGCCTACTTTTGCA
CGCATATTGGCATGATTAAAGCAGTGCAGTCTCGCGGTCCGGTCTCCTGTTTGTCCAGTGACTCGAGACT
CTGCGAAGCTGCACAGATCAGACTGAAGGATCCAAAGGCGTTACTTCTCCAGACTGAAGTTTTACGTGCG
ATATTCTTTCGACAGATCTTAACCTGTTTCCACGGCGGTAATGGTTGTGTTGTACCGTTACCAATCTAAA
AACAGGCAGAGTAAAACAGCGGCAGTCAGATATTGTCTGGCAGGAGGAGCGCGCCTTTCAGACACTTCGA
ATTGCCACAGTTGTAGCATTGTGTTGCGGAGGAACCATTGATTAATTCATATTAACTTCTTTTATAGGTT
TGAGACCGAATAGAAGCACGGACCCTGAATTGCTCGACAAAATGTGTTTAAAATGGCTTTGTTTACACTT
ATAACTAATTTATTGTTAAATAAAACAAATTGTACAAAACCTTTTG
GATTCGACTCTGAGTGAATGATTCATTCACGAAGCTCTATTTAACTGCACGATGTTGCATGGGCTGAGGA
GACATGTTGTTGAAGGCTGCATAAATGTTACTTGTATGAAGTAAGTAGCTTAAGATAGCCTACTTTTGCA
CGCATATTGGCATGATTAAAGCAGTGCAGTCTCGCGGTCCGGTCTCCTGTTTGTCCAGTGACTCGAGACT
CTGCGAAGCTGCACAGATCAGACTGAAGGATCCAAAGGCGTTACTTCTCCAGACTGAAGTTTTACGTGCG
ATATTCTTTCGACAGATCTTAACCTGTTTCCACGGCGGTAATGGTTGTGTTGTACCGTTACCAATCTAAA
AACAGGCAGAGTAAAACAGCGGCAGTCAGATATTGTCTGGCAGGAGGAGCGCGCCTTTCAGACACTTCGA
ATTGCCACAGTTGTAGCATTGTGTTGCGGAGGAACCATTGATTAATTCATATTAACTTCTTTTATAGGTT
TGAGACCGAATAGAAGCACGGACCCTGAATTGCTCGACAAAATGTGTTTAAAATGGCTTTGTTTACACTT
ATAACTAATTTATTGTTAAATAAAACAAATTGTACAAAACCTTTTG