ZFLNCG11020
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035239 | liver | transgenic mCherry control | 183.49 |
| SRR1035237 | liver | transgenic UHRF1 | 18.73 |
| SRR1647679 | head kidney | normal | 14.46 |
| SRR1647682 | spleen | normal | 13.26 |
| SRR527835 | tail | normal | 11.04 |
| SRR1647681 | head kidney | SVCV treatment | 6.70 |
| SRR726542 | 5 dpf | infection with control | 5.71 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 4.48 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 4.22 |
| SRR1291414 | 5 dpi | normal | 4.16 |
Express in tissues
Gene Ontology
Download
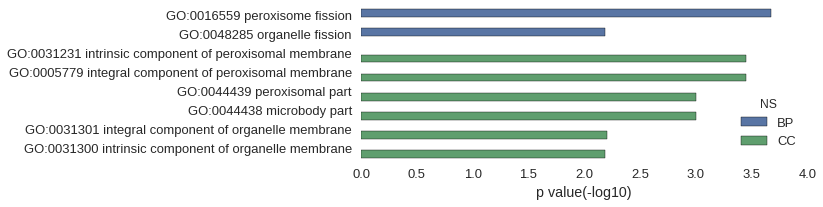
| GO | P value |
|---|---|
| GO:0016559 | 2.13e-04 |
| GO:0048285 | 6.47e-03 |
| GO:0031231 | 3.55e-04 |
| GO:0005779 | 3.55e-04 |
| GO:0044439 | 9.95e-04 |
| GO:0044438 | 9.95e-04 |
| GO:0031301 | 6.25e-03 |
| GO:0031300 | 6.54e-03 |
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT17045 | ENST00000626627; ENST00000417586; ENST00000428786; ENST00000589560; ENST00000593183; ENST00000617009; ENST00000623833; ENST00000621190; NONHSAT192431; NONHSAT087030; NONHSAT192945; NONHSAT061526; NONHSAT061503; NONHSAT192434; NONHSAT192433; NONHSAT212784; NONHSAT192432; | Collinearity with conserved coding gene |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11020
TGTCTGGGGTTTCCCATTCGTTTGGATCATGCAGGACCGAAGACAGGTTTGTTGTAATAGAAGTCCCCTA
ATAAAGAGAGAATGAAATCTACTGGATTTGTTCAGACTTGGCAGAAATAGAATATATATATATATATATA
TATATATATATATATATATATATATATATATATATATATATAGATAGATAGATAGATAGATAGATAGATA
GATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATTAA
TAGAACCTTTGGAATGAAGTAGTTCCCAATTTTTGTGTCTACAGCTGCTTGTTTGGGGAATCCAAGTGGA
ACAACATCCCCAAATCTTTGAATCTCATGTAAAACAGCTTCAGTATAGGGCATATTAACTCTGTCATCCA
AGCAGGGTTGACGTGACTGTCCAATCACTCTGTCTATCTCTGCCTGGACTTTTTCTGACAGACAAATACA
AGTTTGAACCAAAGTATGGACATTTGGACAATATGAGTATTCTTTAACCTTAAAATAAAACCACACTTAC
TCTGTATTTCTGGGAACTTGATCATAAAAAGCAGACCCCAGCGTAATGTAGTCGCACCCGTCTCTGTCCC
AGCCTCAACTATGTCTAGGCAAGAAATGATTAAACTTTCTATGTTAAAGCCAGCTTGAGGGTCACTCTTT
TTCTGGGGGAAAAATATAACAAATGCTTAATATTAAATCATTCAAATTCAAGTCTTTGAAATTCAATTAG
TTACCTTTTCCATCTCAGTCAGGTAGTTGTCGATAAAGTCACGTGGGTTTGCAGGGTCCCAGTCCTC
TGTCTGGGGTTTCCCATTCGTTTGGATCATGCAGGACCGAAGACAGGTTTGTTGTAATAGAAGTCCCCTA
ATAAAGAGAGAATGAAATCTACTGGATTTGTTCAGACTTGGCAGAAATAGAATATATATATATATATATA
TATATATATATATATATATATATATATATATATATATATATAGATAGATAGATAGATAGATAGATAGATA
GATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATAGATTAA
TAGAACCTTTGGAATGAAGTAGTTCCCAATTTTTGTGTCTACAGCTGCTTGTTTGGGGAATCCAAGTGGA
ACAACATCCCCAAATCTTTGAATCTCATGTAAAACAGCTTCAGTATAGGGCATATTAACTCTGTCATCCA
AGCAGGGTTGACGTGACTGTCCAATCACTCTGTCTATCTCTGCCTGGACTTTTTCTGACAGACAAATACA
AGTTTGAACCAAAGTATGGACATTTGGACAATATGAGTATTCTTTAACCTTAAAATAAAACCACACTTAC
TCTGTATTTCTGGGAACTTGATCATAAAAAGCAGACCCCAGCGTAATGTAGTCGCACCCGTCTCTGTCCC
AGCCTCAACTATGTCTAGGCAAGAAATGATTAAACTTTCTATGTTAAAGCCAGCTTGAGGGTCACTCTTT
TTCTGGGGGAAAAATATAACAAATGCTTAATATTAAATCATTCAAATTCAAGTCTTTGAAATTCAATTAG
TTACCTTTTCCATCTCAGTCAGGTAGTTGTCGATAAAGTCACGTGGGTTTGCAGGGTCCCAGTCCTC