ZFLNCG11185
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527834 | head | normal | 21.94 |
| SRR527833 | 5 dpf | normal | 18.46 |
| SRR1565815 | brain | normal | 11.13 |
| SRR1004787 | larvae | impdh1a morpholino | 10.49 |
| SRR1167756 | embryo | mpeg1 morpholino | 10.04 |
| SRR592698 | pineal gland | normal | 9.94 |
| SRR1565819 | brain | normal | 9.45 |
| SRR1004785 | larvae | normal | 9.28 |
| SRR1188148 | embryo | Control PBS 0.5 hpi | 9.13 |
| SRR1565805 | brain | normal | 8.55 |
Express in tissues
Correlated coding gene
Download
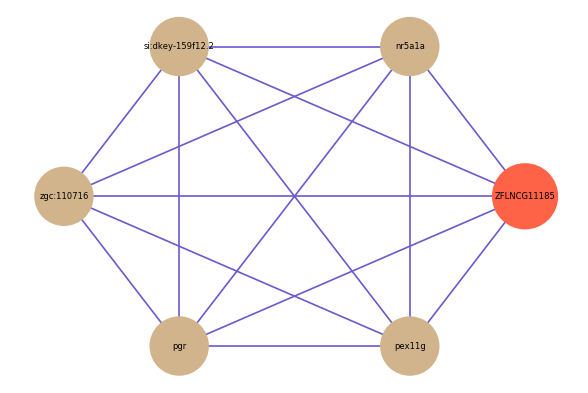
| gene | correlation coefficent |
|---|---|
| nr5a1a | 0.54 |
| si:dkey-159f12.2 | 0.53 |
| zgc:110716 | 0.52 |
| pgr | 0.51 |
| pex11g | 0.51 |
Gene Ontology
Download
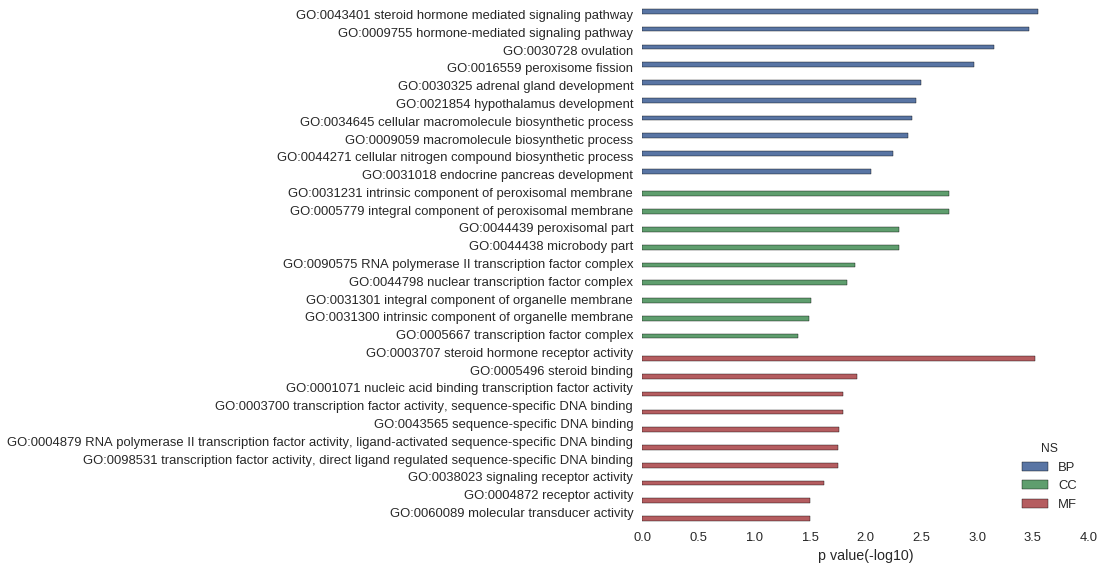
| GO | P value |
|---|---|
| GO:0043401 | 2.85e-04 |
| GO:0009755 | 3.40e-04 |
| GO:0030728 | 7.11e-04 |
| GO:0016559 | 1.07e-03 |
| GO:0030325 | 3.19e-03 |
| GO:0021854 | 3.55e-03 |
| GO:0034645 | 3.84e-03 |
| GO:0009059 | 4.15e-03 |
| GO:0044271 | 5.67e-03 |
| GO:0031018 | 8.85e-03 |
| GO:0031231 | 1.78e-03 |
| GO:0005779 | 1.78e-03 |
| GO:0044439 | 4.97e-03 |
| GO:0044438 | 4.97e-03 |
| GO:0090575 | 1.24e-02 |
| GO:0044798 | 1.45e-02 |
| GO:0031301 | 3.09e-02 |
| GO:0031300 | 3.23e-02 |
| GO:0005667 | 3.99e-02 |
| GO:0003707 | 3.00e-04 |
| GO:0005496 | 1.20e-02 |
| GO:0001071 | 1.58e-02 |
| GO:0003700 | 1.58e-02 |
| GO:0043565 | 1.71e-02 |
| GO:0004879 | 1.76e-02 |
| GO:0098531 | 1.76e-02 |
| GO:0038023 | 2.33e-02 |
| GO:0004872 | 3.15e-02 |
| GO:0060089 | 3.15e-02 |
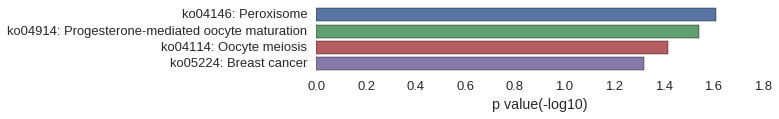
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11185
GATAAAGCGATGGAGTCGAAGCGCCGACGCTTTCTGGAGGAAGGGTCTCAATAGTGGTGACAACATGGTT
CTGGTGCTCGGGCATCATCTCTAAAGCTGAAAACTAAAGCGATCGATCAGTAAAAGCGCTGCTCATTCCT
AAGGCTTTGGTTATTCCGGGCAGCTCCAGCTCTCCATGCCGCAGGTGAGGATATAAACAGGTGGATGGAT
GAATGGCTGGATGTATCTCCGGACAGTGGTGGAGGGAGATGCTGGAGGATCCTATAAGTCGCTGGACGAC
ACTGTTATTATGATCCTTGTCCTCTGTTTGTTTTTTGAGCAGATGTTCGCGGGGGTTTCGGTGCGCATTT
TCTGGACGCCAGAGCAGGACTGCCGTGATTATT
GATAAAGCGATGGAGTCGAAGCGCCGACGCTTTCTGGAGGAAGGGTCTCAATAGTGGTGACAACATGGTT
CTGGTGCTCGGGCATCATCTCTAAAGCTGAAAACTAAAGCGATCGATCAGTAAAAGCGCTGCTCATTCCT
AAGGCTTTGGTTATTCCGGGCAGCTCCAGCTCTCCATGCCGCAGGTGAGGATATAAACAGGTGGATGGAT
GAATGGCTGGATGTATCTCCGGACAGTGGTGGAGGGAGATGCTGGAGGATCCTATAAGTCGCTGGACGAC
ACTGTTATTATGATCCTTGTCCTCTGTTTGTTTTTTGAGCAGATGTTCGCGGGGGTTTCGGTGCGCATTT
TCTGGACGCCAGAGCAGGACTGCCGTGATTATT