ZFLNCG11202
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 66.95 |
| SRR957180 | heart | 7 days after heart tip amputation | 49.34 |
| SRR1188156 | embryo | Control PBS 4 hpi | 41.97 |
| SRR592701 | pineal gland | normal | 38.91 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 31.66 |
| SRR726542 | 5 dpf | infection with control | 31.03 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 28.63 |
| SRR1291417 | 5 dpi | injected marinum | 25.32 |
| SRR941749 | anterior pectoral fin | normal | 25.27 |
| SRR941753 | posterior pectoral fin | normal | 24.95 |
Express in tissues
Correlated coding gene
Download
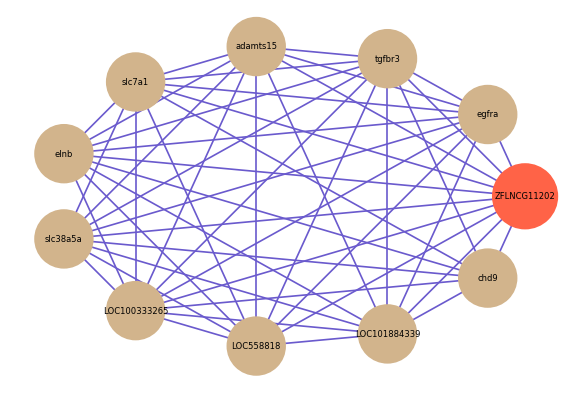
| gene | correlation coefficent |
|---|---|
| egfra | 0.63 |
| tgfbr3 | 0.58 |
| adamts15 | 0.57 |
| slc7a1 | 0.56 |
| elnb | 0.56 |
| slc38a5a | 0.56 |
| chd9 | 0.56 |
| LOC100333265 | 0.56 |
| LOC558818 | 0.56 |
| LOC101884339 | 0.56 |
Gene Ontology
Download
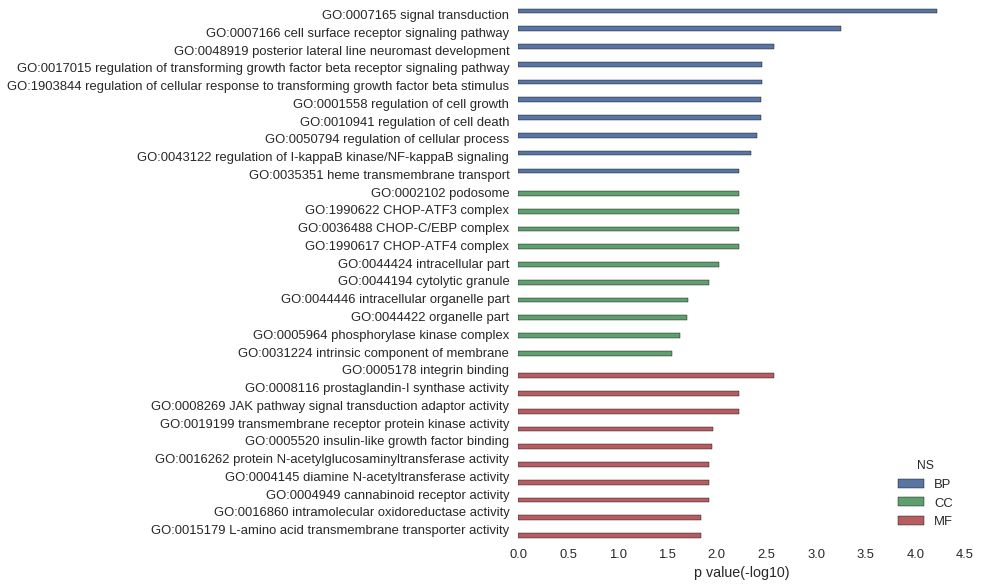
| GO | P value |
|---|---|
| GO:0007165 | 5.97e-05 |
| GO:0007166 | 5.66e-04 |
| GO:0048919 | 2.63e-03 |
| GO:0017015 | 3.52e-03 |
| GO:1903844 | 3.52e-03 |
| GO:0001558 | 3.57e-03 |
| GO:0010941 | 3.60e-03 |
| GO:0050794 | 3.94e-03 |
| GO:0043122 | 4.52e-03 |
| GO:0035351 | 5.97e-03 |
| GO:0002102 | 5.97e-03 |
| GO:1990622 | 5.97e-03 |
| GO:0036488 | 5.97e-03 |
| GO:1990617 | 5.97e-03 |
| GO:0044424 | 9.46e-03 |
| GO:0044194 | 1.19e-02 |
| GO:0044446 | 1.97e-02 |
| GO:0044422 | 1.99e-02 |
| GO:0005964 | 2.37e-02 |
| GO:0031224 | 2.80e-02 |
| GO:0005178 | 2.63e-03 |
| GO:0008116 | 5.97e-03 |
| GO:0008269 | 5.97e-03 |
| GO:0019199 | 1.09e-02 |
| GO:0005520 | 1.12e-02 |
| GO:0016262 | 1.19e-02 |
| GO:0004145 | 1.19e-02 |
| GO:0004949 | 1.19e-02 |
| GO:0016860 | 1.46e-02 |
| GO:0015179 | 1.46e-02 |
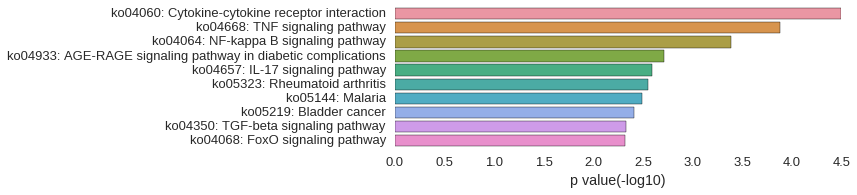
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11202
CACACATATTTGCAAGCAAATCTATCAATATTTCACTGTTGTTTTTAGCAGCACACCACCGTCCAGCAGC
TTCTGTCCTCACAGCAGGATCTACAGAGTTCAGCAGGTTTAGTCAGAATTATAAAGCGCTCCGAGAGGTC
AGAAGTCAGAGGTGGAGGTCAAACCGAGCCGTGACCTCCATCACTGACCGTCACGTGTGCTTGTGTCTG
CACACATATTTGCAAGCAAATCTATCAATATTTCACTGTTGTTTTTAGCAGCACACCACCGTCCAGCAGC
TTCTGTCCTCACAGCAGGATCTACAGAGTTCAGCAGGTTTAGTCAGAATTATAAAGCGCTCCGAGAGGTC
AGAAGTCAGAGGTGGAGGTCAAACCGAGCCGTGACCTCCATCACTGACCGTCACGTGTGCTTGTGTCTG