ZFLNCG11269
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562531 | muscle | normal | 250.64 |
| SRR891495 | heart | normal | 217.52 |
| SRR1562530 | liver | normal | 196.49 |
| SRR1562529 | intestine and pancreas | normal | 138.85 |
| SRR776508 | embryo | miR-146 knockdown | 92.05 |
| SRR527833 | 5 dpf | normal | 91.70 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 85.59 |
| SRR1647681 | head kidney | SVCV treatment | 85.19 |
| SRR1167756 | embryo | mpeg1 morpholino | 81.90 |
| SRR527834 | head | normal | 81.70 |
Express in tissues
Correlated coding gene
Download
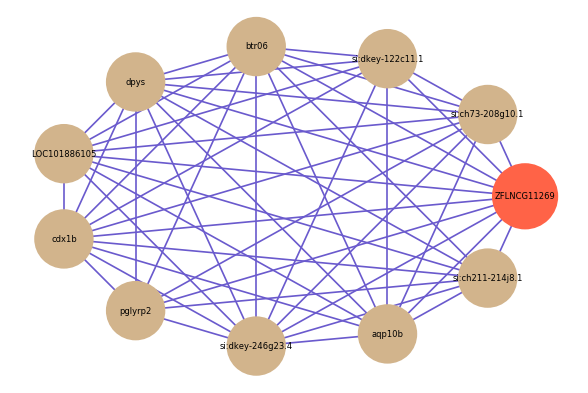
| gene | correlation coefficent |
|---|---|
| si:ch73-208g10.1 | 0.61 |
| si:dkey-122c11.1 | 0.60 |
| si:ch211-214j8.1 | 0.59 |
| btr06 | 0.58 |
| dpys | 0.58 |
| LOC101886105 | 0.57 |
| cdx1b | 0.57 |
| pglyrp2 | 0.57 |
| si:dkey-246g23.4 | 0.57 |
| aqp10b | 0.56 |
Gene Ontology
Download
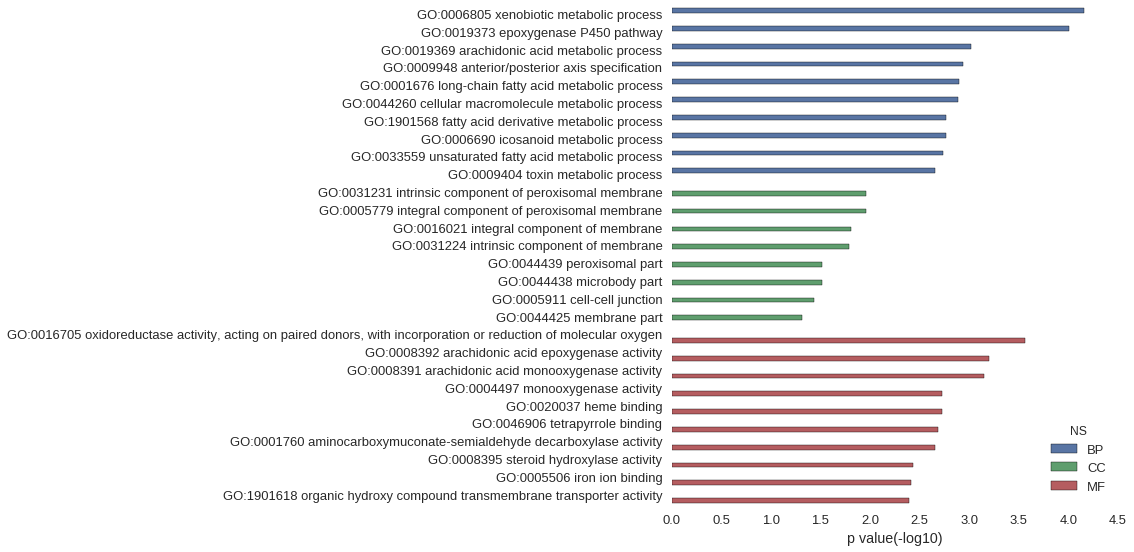
| GO | P value |
|---|---|
| GO:0006805 | 7.01e-05 |
| GO:0019373 | 9.80e-05 |
| GO:0019369 | 9.61e-04 |
| GO:0009948 | 1.15e-03 |
| GO:0001676 | 1.26e-03 |
| GO:0044260 | 1.28e-03 |
| GO:1901568 | 1.71e-03 |
| GO:0006690 | 1.71e-03 |
| GO:0033559 | 1.84e-03 |
| GO:0009404 | 2.20e-03 |
| GO:0031231 | 1.10e-02 |
| GO:0005779 | 1.10e-02 |
| GO:0016021 | 1.55e-02 |
| GO:0031224 | 1.61e-02 |
| GO:0044439 | 3.04e-02 |
| GO:0044438 | 3.04e-02 |
| GO:0005911 | 3.64e-02 |
| GO:0044425 | 4.89e-02 |
| GO:0016705 | 2.74e-04 |
| GO:0008392 | 6.26e-04 |
| GO:0008391 | 7.03e-04 |
| GO:0004497 | 1.88e-03 |
| GO:0020037 | 1.88e-03 |
| GO:0046906 | 2.07e-03 |
| GO:0001760 | 2.20e-03 |
| GO:0008395 | 3.65e-03 |
| GO:0005506 | 3.82e-03 |
| GO:1901618 | 4.01e-03 |
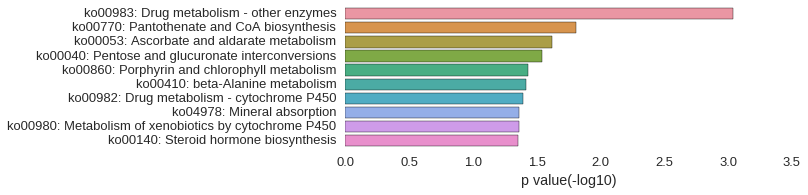
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11269
GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGACCCTTCACCTAGTTCTGTCAGTAGTCTGGAGTCTGG
ACAAGGACAAATACAAAGGCACTAAGCATCTAAACATCTTATTAAGATCATATCATCATTATAAAACAAC
ATTAATCATTACATTACCAAACATGACATTAAAATGACCGTTACGAGACCTGCAAATAACACTTTAACAC
ACAAACAAGTAAACGGATTCATTCTTTCGTCTATATAACAACACCGTTTAAGATATGCATTAGAATTTAG
CATTTACGCAAAATGTTGGTGGCGAAGCAAGTTTATAAATCATATTTTACCTGTTATTGTAGTGAACGTG
GAGAGAGAATAATGTTTGATGTGTCGCCGCGTCGATCTAATGACTACAGCAGTGCATGAGCCGTAGGACC
CAAGTGAGACCCAAGTCAGTGCCTGAGCAGAAAAAAGAGACGCGGTTACATACTTCATTTCTCTTTTAAT
AAAAGTGACATTTCAATTATATGAAGTATTAAAATATACGATATTATAAAATGTCATTATTAGCGTTAAA
ACACTGCAAAACTGTATATTTTGATTTAAAGGCAATACATTGGCAGCCTTAATGGCGGCTGGACGGGATT
GCTC
GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGACCCTTCACCTAGTTCTGTCAGTAGTCTGGAGTCTGG
ACAAGGACAAATACAAAGGCACTAAGCATCTAAACATCTTATTAAGATCATATCATCATTATAAAACAAC
ATTAATCATTACATTACCAAACATGACATTAAAATGACCGTTACGAGACCTGCAAATAACACTTTAACAC
ACAAACAAGTAAACGGATTCATTCTTTCGTCTATATAACAACACCGTTTAAGATATGCATTAGAATTTAG
CATTTACGCAAAATGTTGGTGGCGAAGCAAGTTTATAAATCATATTTTACCTGTTATTGTAGTGAACGTG
GAGAGAGAATAATGTTTGATGTGTCGCCGCGTCGATCTAATGACTACAGCAGTGCATGAGCCGTAGGACC
CAAGTGAGACCCAAGTCAGTGCCTGAGCAGAAAAAAGAGACGCGGTTACATACTTCATTTCTCTTTTAAT
AAAAGTGACATTTCAATTATATGAAGTATTAAAATATACGATATTATAAAATGTCATTATTAGCGTTAAA
ACACTGCAAAACTGTATATTTTGATTTAAAGGCAATACATTGGCAGCCTTAATGGCGGCTGGACGGGATT
GCTC