ZFLNCG11357
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592699 | pineal gland | normal | 6.48 |
| SRR592702 | pineal gland | normal | 2.57 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 1.63 |
| SRR516126 | skin | male and 5 month | 1.43 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 1.41 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 1.38 |
| SRR516124 | skin | male and 3.5 year | 1.38 |
| SRR800045 | muscle | normal | 1.21 |
| SRR516125 | skin | male and 3.5 year | 1.20 |
| SRR658539 | bud | Gata5/6 morphant | 1.15 |
Express in tissues
Correlated coding gene
Download
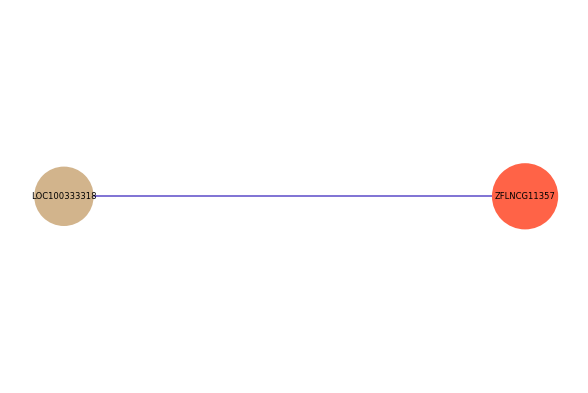
| gene | correlation coefficent |
|---|---|
| LOC100333318 | 0.51 |
Gene Ontology
Gene Ontology of this lncRNA gene hava does not exist!
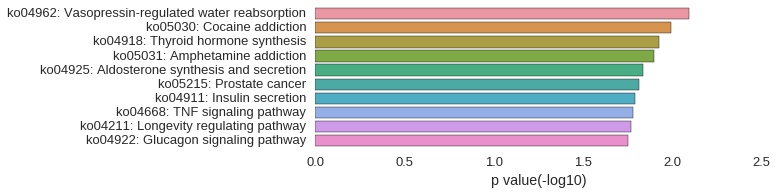
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11357
GCAGAATGAACCGCCAACTTATCCAGCATATGTTTTACCACTGTACAGGACATCATGAACTGATCAAGCT
GGAGGTGGGCGGAGGCCAAATACATTTACTGGAGTGAAACTGGGCGACACGGTGGCTCAGTGGTTTGCAC
TGTGGCCTCACAGCACTCAGGTCACTTGTTCGAGTCTCGGCTGGGTCAGTTGGTGTTTCTGTGTGGAGTT
TGCATGTTCTCCCCGTGTTAGTGTGGGTTTTCTCTGGGTGTGAGATCAACAATGATTAGAAAATGCATAA
TTAATACATCAAGTGAGCAATAAACTGGCCACATGAGCACACAACATGTGTAAAGCACATTTCTCTGGGC
TGTTTCTCCATCTCACCAGACACTGGTCAGGACCCCTGTGTGGAGCCGTGAGGACAGCAGCAGCTGCTGG
ACTGAGTGTATGTGGAGAGTCTGCAGAATCACGACACCACCAGTCTGAAACTCTGTCAGATCATACTGTG
GCCTGTGGAAACCTGCGCTCATCACCTGAGTGTGTCAGTGTGTGCAGGTATAGAAATGACCTGCTCTCTT
CAGTTGTTTGATTGTTTTGACTGTGTGTTCATAATAAA
GCAGAATGAACCGCCAACTTATCCAGCATATGTTTTACCACTGTACAGGACATCATGAACTGATCAAGCT
GGAGGTGGGCGGAGGCCAAATACATTTACTGGAGTGAAACTGGGCGACACGGTGGCTCAGTGGTTTGCAC
TGTGGCCTCACAGCACTCAGGTCACTTGTTCGAGTCTCGGCTGGGTCAGTTGGTGTTTCTGTGTGGAGTT
TGCATGTTCTCCCCGTGTTAGTGTGGGTTTTCTCTGGGTGTGAGATCAACAATGATTAGAAAATGCATAA
TTAATACATCAAGTGAGCAATAAACTGGCCACATGAGCACACAACATGTGTAAAGCACATTTCTCTGGGC
TGTTTCTCCATCTCACCAGACACTGGTCAGGACCCCTGTGTGGAGCCGTGAGGACAGCAGCAGCTGCTGG
ACTGAGTGTATGTGGAGAGTCTGCAGAATCACGACACCACCAGTCTGAAACTCTGTCAGATCATACTGTG
GCCTGTGGAAACCTGCGCTCATCACCTGAGTGTGTCAGTGTGTGCAGGTATAGAAATGACCTGCTCTCTT
CAGTTGTTTGATTGTTTTGACTGTGTGTTCATAATAAA