ZFLNCG11441
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527835 | tail | normal | 18.62 |
| SRR658537 | bud | Gata6 morphant | 14.84 |
| SRR658535 | bud | Gata5 morphant | 12.27 |
| SRR658533 | bud | normal | 11.91 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 11.11 |
| SRR658543 | 6 somite | Gata5 morphan | 11.01 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 10.64 |
| SRR1035978 | 13 hpf | rx3-/- | 10.38 |
| SRR658547 | 6 somite | Gata5/6 morphant | 10.30 |
| SRR658545 | 6 somite | Gata6 morphant | 10.28 |
Express in tissues
Correlated coding gene
Download
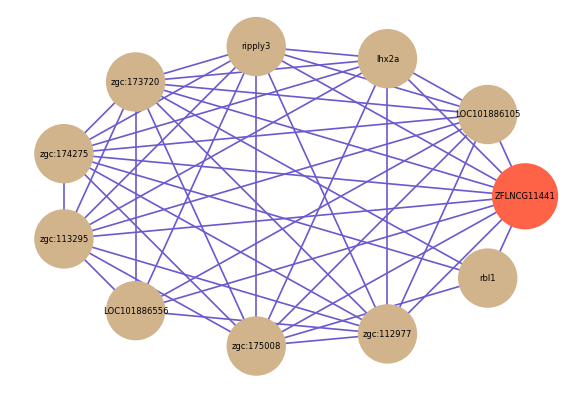
| gene | correlation coefficent |
|---|---|
| LOC101886105 | 0.60 |
| lhx2a | 0.57 |
| ripply3 | 0.57 |
| zgc:173720 | 0.56 |
| zgc:174275 | 0.55 |
| zgc:113295 | 0.54 |
| LOC101886556 | 0.53 |
| rbl1 | 0.52 |
| zgc:175008 | 0.52 |
| zgc:112977 | 0.52 |
Gene Ontology
Download
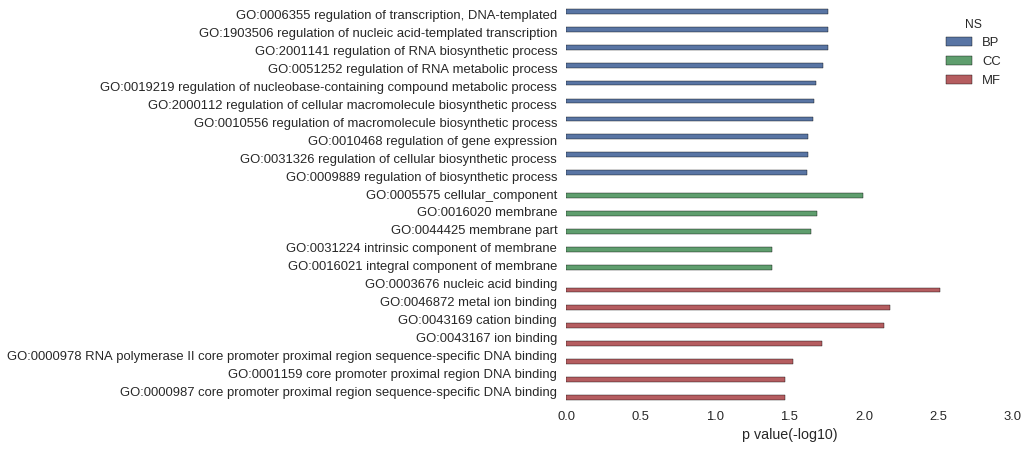
| GO | P value |
|---|---|
| GO:0006355 | 1.74e-02 |
| GO:1903506 | 1.74e-02 |
| GO:2001141 | 1.75e-02 |
| GO:0051252 | 1.87e-02 |
| GO:0019219 | 2.10e-02 |
| GO:2000112 | 2.15e-02 |
| GO:0010556 | 2.20e-02 |
| GO:0010468 | 2.36e-02 |
| GO:0031326 | 2.37e-02 |
| GO:0009889 | 2.40e-02 |
| GO:0005575 | 1.02e-02 |
| GO:0016020 | 2.08e-02 |
| GO:0044425 | 2.26e-02 |
| GO:0031224 | 4.14e-02 |
| GO:0016021 | 4.15e-02 |
| GO:0003676 | 3.10e-03 |
| GO:0046872 | 6.66e-03 |
| GO:0043169 | 7.38e-03 |
| GO:0043167 | 1.90e-02 |
| GO:0000978 | 3.01e-02 |
| GO:0001159 | 3.39e-02 |
| GO:0000987 | 3.39e-02 |
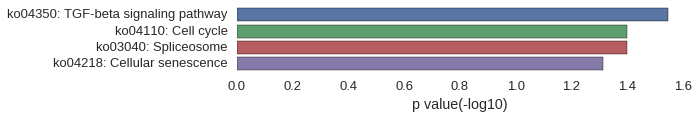
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11441
GATGGGGACAGAGAGAGTGTGGAGAACATTTTAGATGAAGAAAGACAAGCACTTGTCAGTCCTACGTCTC
CCCAATTCCACCATGAAGAAGTAATCATCACATCTGAGCAGAGGTCAACACAAAATGAAGCTAGTTTGAA
TGAAGATGATCTGCCGCAGTTTACTGAAAATGAGGATGTCAACCAGTCAGAAGAGAAGAGGGAGGAGCTA
ATAGATTCAGAGCCTGCCAATCAACCACCCGAGCCTGCAGAGGAGCTTATAGAGTCTCAATCAGCCAATC
AGAGCACTGAGGCTGATGAGGAGAAGGAGCTAATAGACTCTGAACCAACCAATCAGAGCACTAAGCCTGA
GGATGATGAGGAGGAGGAGGAGCTAATGGACTCTGAACCAACCAATCAGAGCACTAAGCCTGATGATGAT
GAGGATGAGGAGGAGCTAATGGACTCTGAACCAACCAATCAAAGCACTAAGCCTGATGATGATGAGGAGG
AGGAGGAGCTAATAGACTCTGAACCAACCAATCAGAGCACTGAACCACCACAGGATGGGGAGGACATGAT
GGAATCCGAATTAGCCAATCAGAGTCTTCAACCTGCAAAGGATAAAGAGGAGATGCTAGAATCAGAGCCA
GCTAATCTGACGTCTGAAGCTGCGGTGGAGGAGGAAGAGGATATTATTGACTCCGAACCAGCCAATCAGA
GCTACGAATTTGCCCAGGAAGAGGAGGAAGAAGAGGATGTTGATG
GATGGGGACAGAGAGAGTGTGGAGAACATTTTAGATGAAGAAAGACAAGCACTTGTCAGTCCTACGTCTC
CCCAATTCCACCATGAAGAAGTAATCATCACATCTGAGCAGAGGTCAACACAAAATGAAGCTAGTTTGAA
TGAAGATGATCTGCCGCAGTTTACTGAAAATGAGGATGTCAACCAGTCAGAAGAGAAGAGGGAGGAGCTA
ATAGATTCAGAGCCTGCCAATCAACCACCCGAGCCTGCAGAGGAGCTTATAGAGTCTCAATCAGCCAATC
AGAGCACTGAGGCTGATGAGGAGAAGGAGCTAATAGACTCTGAACCAACCAATCAGAGCACTAAGCCTGA
GGATGATGAGGAGGAGGAGGAGCTAATGGACTCTGAACCAACCAATCAGAGCACTAAGCCTGATGATGAT
GAGGATGAGGAGGAGCTAATGGACTCTGAACCAACCAATCAAAGCACTAAGCCTGATGATGATGAGGAGG
AGGAGGAGCTAATAGACTCTGAACCAACCAATCAGAGCACTGAACCACCACAGGATGGGGAGGACATGAT
GGAATCCGAATTAGCCAATCAGAGTCTTCAACCTGCAAAGGATAAAGAGGAGATGCTAGAATCAGAGCCA
GCTAATCTGACGTCTGAAGCTGCGGTGGAGGAGGAAGAGGATATTATTGACTCCGAACCAGCCAATCAGA
GCTACGAATTTGCCCAGGAAGAGGAGGAAGAAGAGGATGTTGATG