ZFLNCG11875
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647679 | head kidney | normal | 94.97 |
| SRR1647680 | head kidney | SVCV treatment | 87.17 |
| SRR1647681 | head kidney | SVCV treatment | 63.07 |
| SRR658539 | bud | Gata5/6 morphant | 52.76 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 43.16 |
| SRR1188156 | embryo | Control PBS 4 hpi | 36.77 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 35.20 |
| SRR1188160 | embryo | Insulin 4 hpi | 22.54 |
| SRR1291417 | 5 dpi | injected marinum | 22.26 |
| SRR1188148 | embryo | Control PBS 0.5 hpi | 19.80 |
Express in tissues
Correlated coding gene
Download
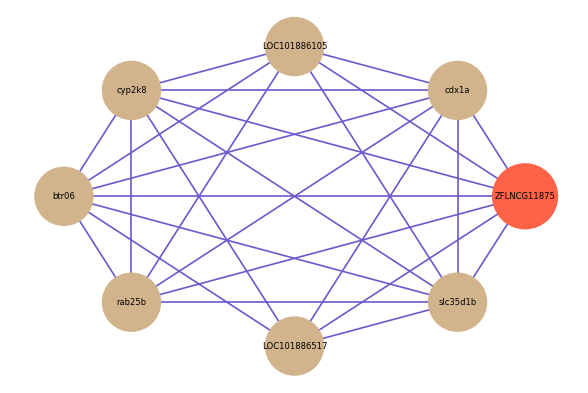
| gene | correlation coefficent |
|---|---|
| cdx1a | 0.57 |
| LOC101886105 | 0.55 |
| cyp2k8 | 0.53 |
| btr06 | 0.52 |
| rab25b | 0.52 |
| LOC101886517 | 0.52 |
| slc35d1b | 0.52 |
Gene Ontology
Download
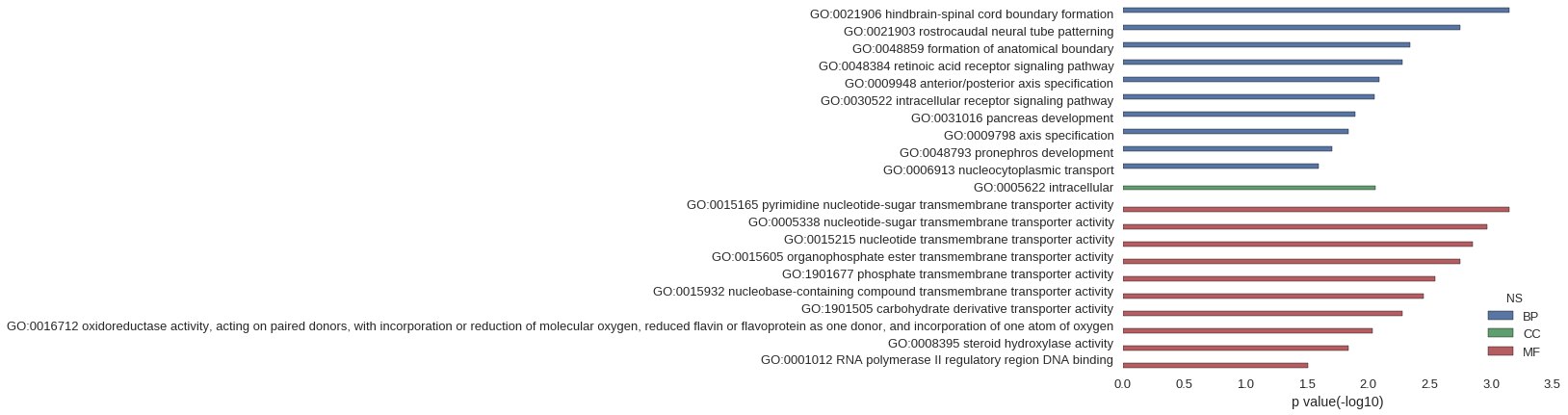
| GO | P value |
|---|---|
| GO:0021906 | 7.11e-04 |
| GO:0021903 | 1.78e-03 |
| GO:0048859 | 4.61e-03 |
| GO:0048384 | 5.32e-03 |
| GO:0009948 | 8.15e-03 |
| GO:0030522 | 8.85e-03 |
| GO:0031016 | 1.27e-02 |
| GO:0009798 | 1.45e-02 |
| GO:0048793 | 1.97e-02 |
| GO:0006913 | 2.53e-02 |
| GO:0005622 | 8.80e-03 |
| GO:0015165 | 7.11e-04 |
| GO:0005338 | 1.07e-03 |
| GO:0015215 | 1.42e-03 |
| GO:0015605 | 1.78e-03 |
| GO:1901677 | 2.84e-03 |
| GO:0015932 | 3.55e-03 |
| GO:1901505 | 5.32e-03 |
| GO:0016712 | 9.21e-03 |
| GO:0008395 | 1.45e-02 |
| GO:0001012 | 3.09e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11875
GTCGCGACTCCACCCACTTCCAGAAACAACTCCACCCATACTAAAGTAGTGGCCCCACCTCCTCAAACAA
AAACTCCACCTCACAAACAGACCACACCCCCTCGTACAGAAGAACCGCCCGTCAGAGAGGCCCCGCCCCC
TCAGAAGCCATATACGGAGCGTGAGAGGAACGTGGTGAGTGTTTGACAGTTCAGTTAATAATATCCATAA
TTAACATCAATCAATAATTGGTGCGAGTTTTCGTTAATGTAAATGCTTTTTGACCAACGATTTTACTGTT
AGTGTTCGTTATTGTTGACTCAAATAACCTTGATTTAAACATTCAAGTGTGGATGTGAAATGAAGATGCT
CATTCCTCTGCAGGACACAATCTTCATAGTGAAAAGTGCTGTAGAAATAAAGATGAGTTGAATTGAATTG
CAGGATATCCTGAATCACCTTATTAACGACATCGAAGCGTTCGTTGGCAGACTGACG
GTCGCGACTCCACCCACTTCCAGAAACAACTCCACCCATACTAAAGTAGTGGCCCCACCTCCTCAAACAA
AAACTCCACCTCACAAACAGACCACACCCCCTCGTACAGAAGAACCGCCCGTCAGAGAGGCCCCGCCCCC
TCAGAAGCCATATACGGAGCGTGAGAGGAACGTGGTGAGTGTTTGACAGTTCAGTTAATAATATCCATAA
TTAACATCAATCAATAATTGGTGCGAGTTTTCGTTAATGTAAATGCTTTTTGACCAACGATTTTACTGTT
AGTGTTCGTTATTGTTGACTCAAATAACCTTGATTTAAACATTCAAGTGTGGATGTGAAATGAAGATGCT
CATTCCTCTGCAGGACACAATCTTCATAGTGAAAAGTGCTGTAGAAATAAAGATGAGTTGAATTGAATTG
CAGGATATCCTGAATCACCTTATTAACGACATCGAAGCGTTCGTTGGCAGACTGACG
