ZFLNCG11958
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 1978.53 |
| ERR023143 | swim bladder | normal | 724.93 |
| ERR023146 | head kidney | normal | 217.14 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 114.01 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 112.77 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 107.97 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 97.24 |
| SRR1299125 | caudal fin | Half day time after treatment | 81.89 |
| ERR023144 | brain | normal | 81.86 |
| SRR1299127 | caudal fin | Two days time after treatment | 76.97 |
Express in tissues
Correlated coding gene
Download
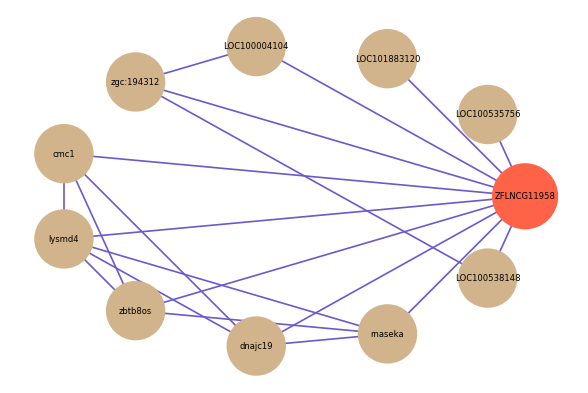
| gene | correlation coefficent |
|---|---|
| LOC100535756 | 0.54 |
| LOC101883120 | 0.54 |
| LOC100004104 | 0.52 |
| cmc1 | 0.52 |
| lysmd4 | 0.51 |
| zbtb8os | 0.51 |
| LOC100538148 | 0.51 |
| rnaseka | 0.51 |
| dnajc19 | 0.50 |
| zgc:194312 | 0.50 |
Gene Ontology
Download
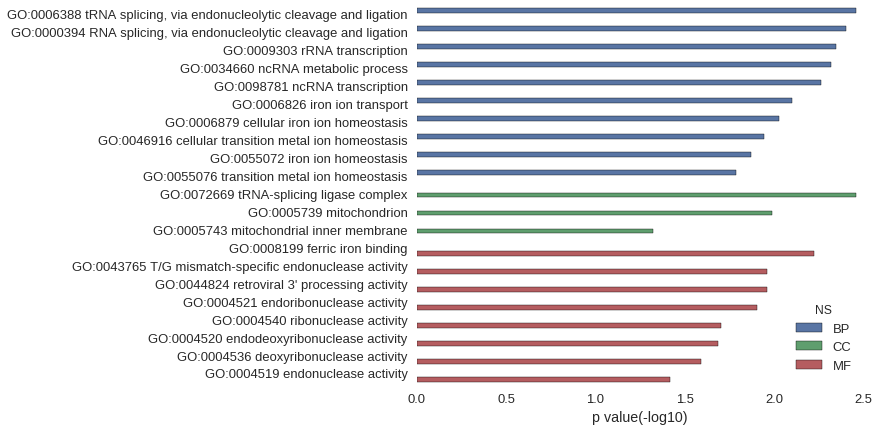
| GO | P value |
|---|---|
| GO:0006388 | 3.48e-03 |
| GO:0000394 | 3.97e-03 |
| GO:0009303 | 4.47e-03 |
| GO:0034660 | 4.81e-03 |
| GO:0098781 | 5.46e-03 |
| GO:0006826 | 7.93e-03 |
| GO:0006879 | 9.42e-03 |
| GO:0046916 | 1.14e-02 |
| GO:0055072 | 1.34e-02 |
| GO:0055076 | 1.63e-02 |
| GO:0072669 | 3.48e-03 |
| GO:0005739 | 1.03e-02 |
| GO:0005743 | 4.78e-02 |
| GO:0008199 | 5.96e-03 |
| GO:0043765 | 1.09e-02 |
| GO:0044824 | 1.09e-02 |
| GO:0004521 | 1.24e-02 |
| GO:0004540 | 1.97e-02 |
| GO:0004520 | 2.07e-02 |
| GO:0004536 | 2.56e-02 |
| GO:0004519 | 3.82e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11958
CACAATTGGCAGTCACAAAAACAAACGAAAAGACTGAGGAACACAAATGACAATACAATTGAGTGCATAT
ACATAAACGTATGCTGAATTATTCTGTATTTAAAACACAAAAACTGCTTTATTTTCCATAGATTCTGCCA
ATTTTCATCTTAGATGTACAGTATTTGTGACCATGGTCTTTGTAGTGACCACAAGCGTACATTAATCATA
TCAGATTTATGTATCATCTAGAAGCATTTTTGGCTTGATTGATGGTTTGATAGGGTGGGATATTTGGCAG
AGCTACAACAGTTTGAAAATATTTATCAAAAAATTA
CACAATTGGCAGTCACAAAAACAAACGAAAAGACTGAGGAACACAAATGACAATACAATTGAGTGCATAT
ACATAAACGTATGCTGAATTATTCTGTATTTAAAACACAAAAACTGCTTTATTTTCCATAGATTCTGCCA
ATTTTCATCTTAGATGTACAGTATTTGTGACCATGGTCTTTGTAGTGACCACAAGCGTACATTAATCATA
TCAGATTTATGTATCATCTAGAAGCATTTTTGGCTTGATTGATGGTTTGATAGGGTGGGATATTTGGCAG
AGCTACAACAGTTTGAAAATATTTATCAAAAAATTA

