ZFLNCG11988
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023146 | head kidney | normal | 14.73 |
| ERR023145 | heart | normal | 11.94 |
| ERR023144 | brain | normal | 10.35 |
| ERR023143 | swim bladder | normal | 7.48 |
| SRR1048061 | pineal gland | dark | 5.63 |
| SRR891511 | brain | normal | 5.09 |
| SRR1647679 | head kidney | normal | 4.45 |
| SRR941749 | anterior pectoral fin | normal | 4.29 |
| SRR592701 | pineal gland | normal | 3.67 |
| SRR516127 | skin | male and 5 month | 3.61 |
Express in tissues
Correlated coding gene
Download
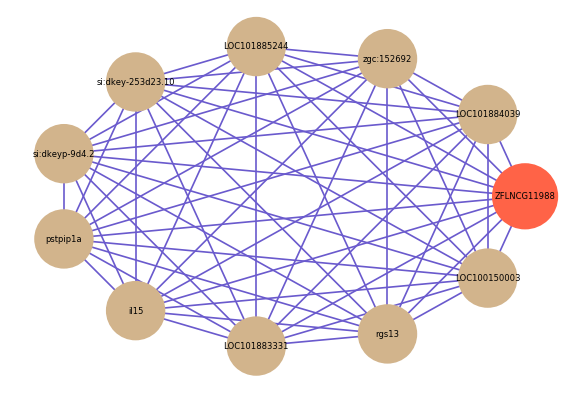
| gene | correlation coefficent |
|---|---|
| LOC101884039 | 0.56 |
| zgc:152692 | 0.56 |
| LOC101885244 | 0.55 |
| si:dkey-253d23.10 | 0.55 |
| si:dkeyp-9d4.2 | 0.55 |
| pstpip1a | 0.55 |
| il15 | 0.54 |
| LOC101883331 | 0.54 |
| rgs13 | 0.54 |
| LOC100150003 | 0.54 |
Gene Ontology
Download
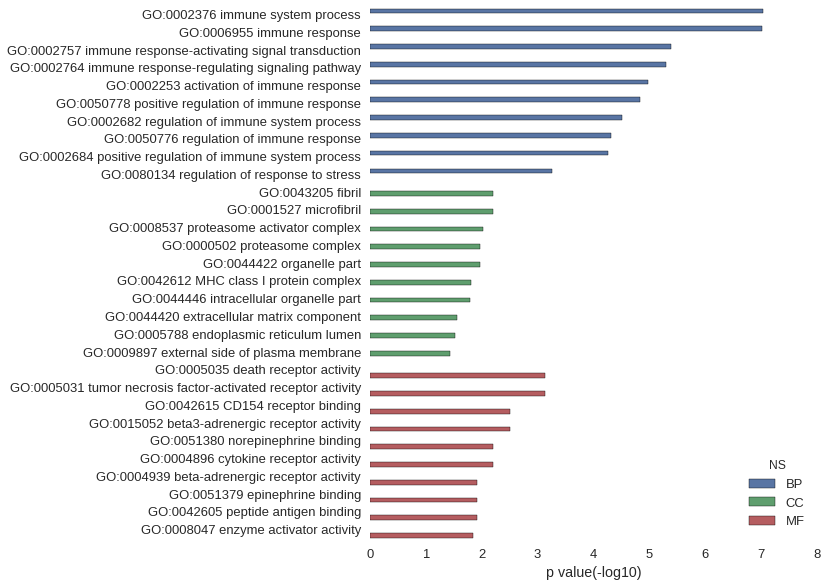
| GO | P value |
|---|---|
| GO:0002376 | 9.10e-08 |
| GO:0006955 | 9.46e-08 |
| GO:0002757 | 4.06e-06 |
| GO:0002764 | 5.09e-06 |
| GO:0002253 | 1.03e-05 |
| GO:0050778 | 1.46e-05 |
| GO:0002682 | 3.06e-05 |
| GO:0050776 | 4.90e-05 |
| GO:0002684 | 5.52e-05 |
| GO:0080134 | 5.41e-04 |
| GO:0043205 | 6.24e-03 |
| GO:0001527 | 6.24e-03 |
| GO:0008537 | 9.35e-03 |
| GO:0000502 | 1.06e-02 |
| GO:0044422 | 1.07e-02 |
| GO:0042612 | 1.55e-02 |
| GO:0044446 | 1.65e-02 |
| GO:0044420 | 2.78e-02 |
| GO:0005788 | 3.08e-02 |
| GO:0009897 | 3.69e-02 |
| GO:0005035 | 7.29e-04 |
| GO:0005031 | 7.29e-04 |
| GO:0042615 | 3.13e-03 |
| GO:0015052 | 3.13e-03 |
| GO:0051380 | 6.24e-03 |
| GO:0004896 | 6.25e-03 |
| GO:0004939 | 1.24e-02 |
| GO:0051379 | 1.24e-02 |
| GO:0042605 | 1.24e-02 |
| GO:0008047 | 1.43e-02 |
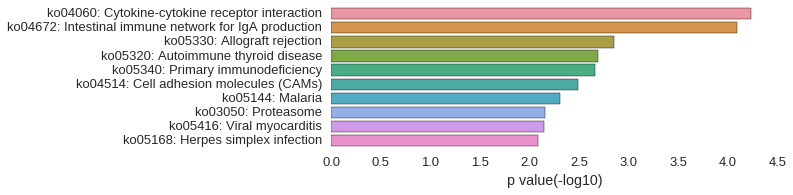
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11988
GATGATGTAAAATAGTCTACTGTAACATCTGTGATTATATAAAATACAAAACATATGAAAACATATTGAG
CCTTACAGTCTCTGAAATGTTTCCAATTACAAAAATACCCAAAACATACATTTAGGCCTTATTTGGGTTC
ATAAACGATTCCAGCATCTTTGTGGGCATGTTTTGACAGGCAGCGATTTCACAGACACACACACACAAAG
AGGATAAAATGATAAACACAGACACACACACACACTAAGAGGATAACATGATAAACACAACAAGTGTGTG
CATGAGGGGGCTGGCTG
GATGATGTAAAATAGTCTACTGTAACATCTGTGATTATATAAAATACAAAACATATGAAAACATATTGAG
CCTTACAGTCTCTGAAATGTTTCCAATTACAAAAATACCCAAAACATACATTTAGGCCTTATTTGGGTTC
ATAAACGATTCCAGCATCTTTGTGGGCATGTTTTGACAGGCAGCGATTTCACAGACACACACACACAAAG
AGGATAAAATGATAAACACAGACACACACACACACTAAGAGGATAACATGATAAACACAACAAGTGTGTG
CATGAGGGGGCTGGCTG