ZFLNCG12163
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800037 | egg | normal | 21.99 |
| SRR700534 | heart | control morpholino | 19.39 |
| SRR1647684 | spleen | SVCV treatment | 17.69 |
| SRR941749 | anterior pectoral fin | normal | 13.58 |
| SRR941753 | posterior pectoral fin | normal | 12.28 |
| SRR1647683 | spleen | SVCV treatment | 9.15 |
| SRR527834 | head | normal | 5.18 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 4.64 |
| SRR516125 | skin | male and 3.5 year | 4.31 |
| SRR1299125 | caudal fin | Half day time after treatment | 4.05 |
Express in tissues
Correlated coding gene
Download
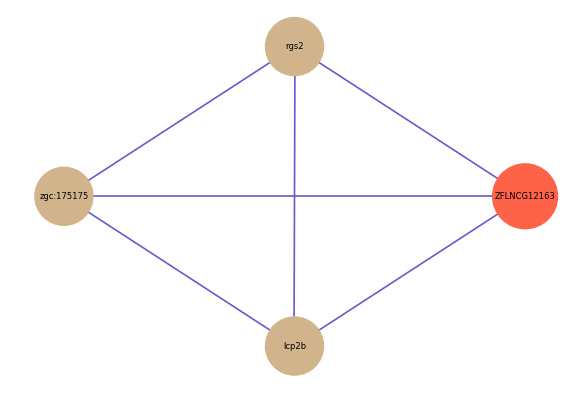
| gene | correlation coefficent |
|---|---|
| rgs2 | 0.60 |
| zgc:175175 | 0.53 |
| lcp2b | 0.52 |
Gene Ontology
Download
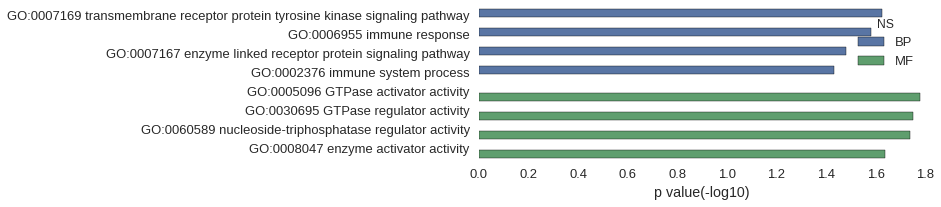
| GO | P value |
|---|---|
| GO:0007169 | 2.36e-02 |
| GO:0006955 | 2.63e-02 |
| GO:0007167 | 3.31e-02 |
| GO:0002376 | 3.68e-02 |
| GO:0005096 | 1.66e-02 |
| GO:0030695 | 1.78e-02 |
| GO:0060589 | 1.83e-02 |
| GO:0008047 | 2.30e-02 |
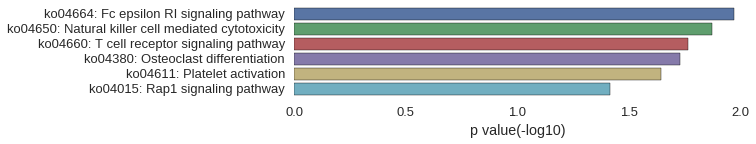
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12163
TCAGCTCAATTTGATTGATAGCAATGGACAGATTGACCAAAATGAGAACTGCCAGTCCAGATTCAGGCAG
GGAGCATTTGATCAACTATAGGAACTCTGAGGAGATGACGGAACAAAGCCAGATAAAGTAAGATGTGCTG
TATGTTGCGTAGTATTTGGTTTGATGTTGATGTCAATATGCGTGTGGTTGCTGATGTTTGTTGATTGCTT
TTAATAGAAAGAAAAGCTGGAGGCCGAGAATATTCTTCAGCACTTCACCTTTAAAGGAATCACAGAACAC
CACGCCACAGAAAAAATCACACTTGTGAGTTGATGGATTCCTTCATATTTTTGTATTTATATTTCATAAT
TTAGCAGACGCTTTCATCTGAATTAAGGCAATCTGTTTGCGTTTACATCAGAAGAAATGTGGAATGTATA
AACAGATGGGATATGATAAATGATGAATGTAATAAGTGTAACATAATTTGCAGAAAATGTCCAAATAAAG
TGTATGTGTGTCATTTTATTTTGATTTTAATCTCTGCGTTTATGTTCCCTCA
TCAGCTCAATTTGATTGATAGCAATGGACAGATTGACCAAAATGAGAACTGCCAGTCCAGATTCAGGCAG
GGAGCATTTGATCAACTATAGGAACTCTGAGGAGATGACGGAACAAAGCCAGATAAAGTAAGATGTGCTG
TATGTTGCGTAGTATTTGGTTTGATGTTGATGTCAATATGCGTGTGGTTGCTGATGTTTGTTGATTGCTT
TTAATAGAAAGAAAAGCTGGAGGCCGAGAATATTCTTCAGCACTTCACCTTTAAAGGAATCACAGAACAC
CACGCCACAGAAAAAATCACACTTGTGAGTTGATGGATTCCTTCATATTTTTGTATTTATATTTCATAAT
TTAGCAGACGCTTTCATCTGAATTAAGGCAATCTGTTTGCGTTTACATCAGAAGAAATGTGGAATGTATA
AACAGATGGGATATGATAAATGATGAATGTAATAAGTGTAACATAATTTGCAGAAAATGTCCAAATAAAG
TGTATGTGTGTCATTTTATTTTGATTTTAATCTCTGCGTTTATGTTCCCTCA