ZFLNCG12187
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748488 | sperm | normal | 0.69 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 0.66 |
| SRR1049814 | 256 cell | normal | 0.57 |
| SRR372787 | 2-4 cell | normal | 0.46 |
| SRR800046 | sphere stage | 5azaCyD treatment | 0.29 |
| ERR023143 | swim bladder | normal | 0.16 |
| SRR372789 | 1K cell | normal | 0.16 |
| SRR1621206 | embryo | normal | 0.16 |
| SRR372793 | shield | normal | 0.11 |
| SRR516127 | skin | male and 5 month | 0.08 |
Express in tissues
Correlated coding gene
Download
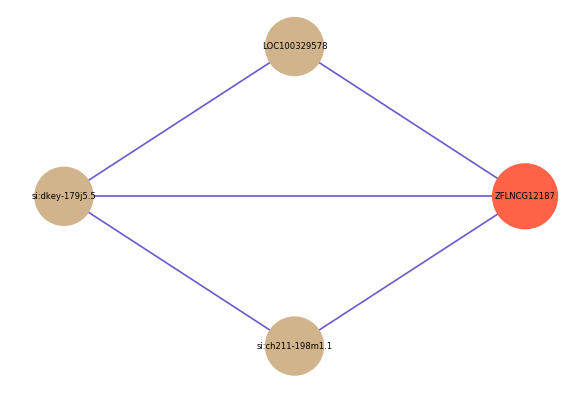
| gene | correlation coefficent |
|---|---|
| LOC100329578 | 0.52 |
| si:ch211-198m1.1 | 0.50 |
| si:dkey-179j5.5 | 0.50 |
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0030246 | 7.11e-03 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12187
ATGAATCAGTAAAGACGACATGCACAGTGGCAGATGACAGCAGAATCATGGTTTCCTCTCAGTTTAGTGC
TCACCTGTGAGTTGCTTTTTGGGTTTGACAAGTTGATGTTCAGTGAATATACCTTTGGCTGCACTGTTGT
GTTGTGAATAGTTTCCCTGTGGTTCAAGTTTTCTGTTAATTATAATGTGAATATGAATAGCAGTAAACGG
GTGTCTTGATCAGTTCTTCTGTGTTTGTTTTTCAGTGATTTTACTGAACTCAAGCCTGCTGATATCAGCG
AGTGGTGGACATTATGGTCACGGACATGGTTACGGTGGACATGATCATGATGACGATCATGACTATGACT
ACTGCAGGAGGTCCTCTAGTAAGAGTTTGTTATTCTAAGACACCTAAATTTCGTTTCATGAACCGAGTGA
TGCTTGACATTTATTTCTTTGCTCCATTAATCCTCTAAAGTGCAACCCTTTATTTCTCAGAAACTATAAA
TGCATGTGAAGGCTCTATCCTGCGTCTGTCCTGTCCTGGTAAGATGAAGTCGCAATCCATTAAATAAGTG
TTTTTCAAATTCTGCTCTTTTAAATGTTACATGGGCTTCTCTCTTGTCTCAACTTTTGGCTTAATTTTTC
TTGCAGGTCGCAGGAGCTGATCATGTTGTTTTTGTCTGATTTTTCTGCCAGTGTCTCCCCTACATTGACA
TTACACTTTTAATAAATGCTTAATGCAAATAA
ATGAATCAGTAAAGACGACATGCACAGTGGCAGATGACAGCAGAATCATGGTTTCCTCTCAGTTTAGTGC
TCACCTGTGAGTTGCTTTTTGGGTTTGACAAGTTGATGTTCAGTGAATATACCTTTGGCTGCACTGTTGT
GTTGTGAATAGTTTCCCTGTGGTTCAAGTTTTCTGTTAATTATAATGTGAATATGAATAGCAGTAAACGG
GTGTCTTGATCAGTTCTTCTGTGTTTGTTTTTCAGTGATTTTACTGAACTCAAGCCTGCTGATATCAGCG
AGTGGTGGACATTATGGTCACGGACATGGTTACGGTGGACATGATCATGATGACGATCATGACTATGACT
ACTGCAGGAGGTCCTCTAGTAAGAGTTTGTTATTCTAAGACACCTAAATTTCGTTTCATGAACCGAGTGA
TGCTTGACATTTATTTCTTTGCTCCATTAATCCTCTAAAGTGCAACCCTTTATTTCTCAGAAACTATAAA
TGCATGTGAAGGCTCTATCCTGCGTCTGTCCTGTCCTGGTAAGATGAAGTCGCAATCCATTAAATAAGTG
TTTTTCAAATTCTGCTCTTTTAAATGTTACATGGGCTTCTCTCTTGTCTCAACTTTTGGCTTAATTTTTC
TTGCAGGTCGCAGGAGCTGATCATGTTGTTTTTGTCTGATTTTTCTGCCAGTGTCTCCCCTACATTGACA
TTACACTTTTAATAAATGCTTAATGCAAATAA
