ZFLNCG12245
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562531 | muscle | normal | 0.73 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 0.43 |
| SRR516135 | skin | male and 5 month | 0.36 |
| SRR535890 | larvae | normal | 0.33 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 0.31 |
| SRR1004789 | larvae | impdh2 morpholino | 0.30 |
| SRR535943 | larvae | vangl mut | 0.30 |
| SRR1167763 | embryo | ptpn6 morpholino and bacterial infection | 0.28 |
| SRR1167757 | embryo | mpeg1 morpholino and bacterial infection | 0.27 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 0.26 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| LOC100535347 | 0.53 |
| si:ch211-136a13.1 | 0.51 |
Gene Ontology
Download
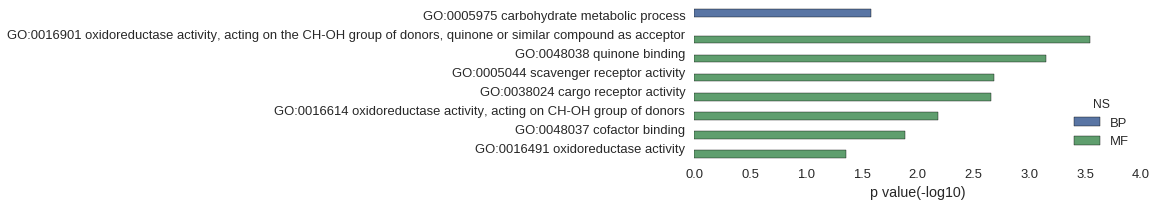
| GO | P value |
|---|---|
| GO:0005975 | 2.62e-02 |
| GO:0016901 | 2.84e-04 |
| GO:0048038 | 7.11e-04 |
| GO:0005044 | 2.06e-03 |
| GO:0038024 | 2.20e-03 |
| GO:0016614 | 6.54e-03 |
| GO:0048037 | 1.29e-02 |
| GO:0016491 | 4.39e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12245
TCTTTTTTTCCTGTTCATCTAAGTCCTGCTATAGCCACTCCTCATTTTATTTGGGCTTGGAACCGGAACA
TGGAATTTCAAATAAACTCCTGTGGCAGAGTTCTGAAACACAAACATCATATGTGTTTAGTTAGTGTGAT
CTACTCATGACATTTACTTCTTGCTACTGTTAAATCTGAATAAACTGAACTTACAGTAAAGAGAGAAATA
ATTCCAATACATACAGCAAACATTATTGCAGTCCTCTTTGGTCTTGGTTTTGTTCCTTGGTTTGTACTTC
CTCTTACTTGTTTTTCTGAATTGACAAAAATGAATAGGAAAAACTATATGAAAACATACATGCACCCATT
CCTAGAGCAAAATATTAATGCGGGGCAAAGATTAATCGCAACCAAAATAAAAGTTGTGTGTGTACTGTAT
ATATGTATTATAAAAATATAAATGCAAACATGCATGCATATGATACAGATTCTAAATATATTCAAATATT
TATGCAATAAAAATGTTTTTACACACCTTTGACAATAAGATTGAAACTCTTTAATATTGATTGTCTTATA
ATATAGCATGTGTATCTTCCTTTATCTCTGTGTTGAAGATTGTTGAGTTTGAGCGAGAAGTTTCCATTCT
TATATTCATCAGGGAAGGTTTCAGCTCTGTTCTTATATTCTTGATTTTGTCCCTCTACTGAAACTTGTCC
ATTAATAATGTCGTACACGTTCAGATTATTGTGTCTCCAGCGCACTATGATGTCTTGAACTGTGAGCTCA
GGTTGTCTAGTAGAGCATTCGATAACAACTGAGTCACCAATAAAACCCACAATGCTCTCCTGCAGAGACA
CTAAAAAGAGAAAAGTTTATTATATAAAAAGAACTAATAAGGCATTTTCTAACTTGACTGTTAAACTTGT
GCTCACTTTCAGTCATCAG
TCTTTTTTTCCTGTTCATCTAAGTCCTGCTATAGCCACTCCTCATTTTATTTGGGCTTGGAACCGGAACA
TGGAATTTCAAATAAACTCCTGTGGCAGAGTTCTGAAACACAAACATCATATGTGTTTAGTTAGTGTGAT
CTACTCATGACATTTACTTCTTGCTACTGTTAAATCTGAATAAACTGAACTTACAGTAAAGAGAGAAATA
ATTCCAATACATACAGCAAACATTATTGCAGTCCTCTTTGGTCTTGGTTTTGTTCCTTGGTTTGTACTTC
CTCTTACTTGTTTTTCTGAATTGACAAAAATGAATAGGAAAAACTATATGAAAACATACATGCACCCATT
CCTAGAGCAAAATATTAATGCGGGGCAAAGATTAATCGCAACCAAAATAAAAGTTGTGTGTGTACTGTAT
ATATGTATTATAAAAATATAAATGCAAACATGCATGCATATGATACAGATTCTAAATATATTCAAATATT
TATGCAATAAAAATGTTTTTACACACCTTTGACAATAAGATTGAAACTCTTTAATATTGATTGTCTTATA
ATATAGCATGTGTATCTTCCTTTATCTCTGTGTTGAAGATTGTTGAGTTTGAGCGAGAAGTTTCCATTCT
TATATTCATCAGGGAAGGTTTCAGCTCTGTTCTTATATTCTTGATTTTGTCCCTCTACTGAAACTTGTCC
ATTAATAATGTCGTACACGTTCAGATTATTGTGTCTCCAGCGCACTATGATGTCTTGAACTGTGAGCTCA
GGTTGTCTAGTAGAGCATTCGATAACAACTGAGTCACCAATAAAACCCACAATGCTCTCCTGCAGAGACA
CTAAAAAGAGAAAAGTTTATTATATAAAAAGAACTAATAAGGCATTTTCTAACTTGACTGTTAAACTTGT
GCTCACTTTCAGTCATCAG
