ZFLNCG12265
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800045 | muscle | normal | 260.05 |
| SRR1004789 | larvae | impdh2 morpholino | 129.44 |
| SRR1004787 | larvae | impdh1a morpholino | 119.14 |
| SRR1035978 | 13 hpf | rx3-/- | 104.87 |
| SRR1569495 | embryo | normal | 104.18 |
| SRR658541 | 6 somite | normal | 101.36 |
| SRR658543 | 6 somite | Gata5 morphan | 100.38 |
| SRR749514 | 24 hpf | normal | 98.08 |
| SRR658545 | 6 somite | Gata6 morphant | 93.49 |
| SRR797911 | embryo | prdm1-/- | 88.43 |
Express in tissues
Correlated coding gene
Download
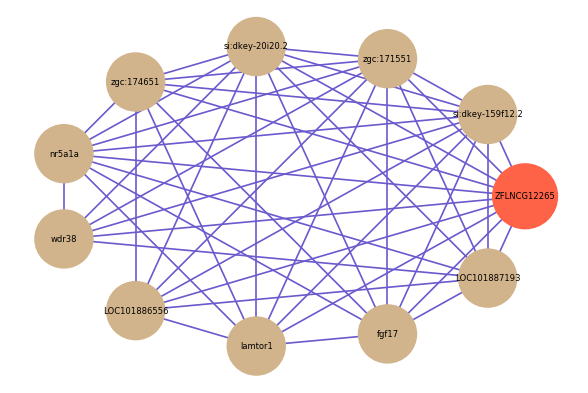
| gene | correlation coefficent |
|---|---|
| si:dkey-159f12.2 | 0.69 |
| zgc:171551 | 0.68 |
| si:dkey-20i20.2 | 0.65 |
| zgc:174651 | 0.63 |
| nr5a1a | 0.61 |
| wdr38 | 0.61 |
| LOC101886556 | 0.60 |
| lamtor1 | 0.60 |
| fgf17 | 0.59 |
| LOC101887193 | 0.59 |
Gene Ontology
Download
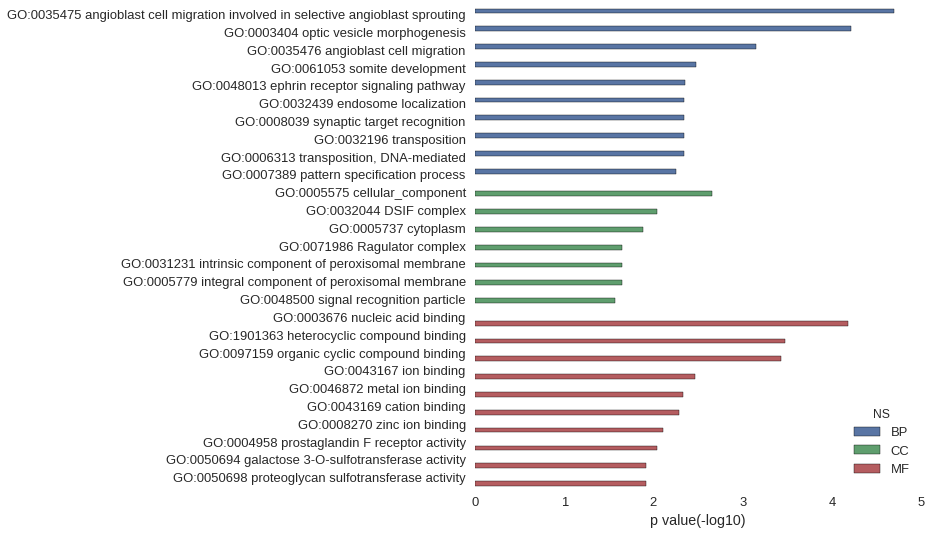
| GO | P value |
|---|---|
| GO:0035475 | 2.04e-05 |
| GO:0003404 | 6.09e-05 |
| GO:0035476 | 7.18e-04 |
| GO:0061053 | 3.31e-03 |
| GO:0048013 | 4.44e-03 |
| GO:0032439 | 4.55e-03 |
| GO:0008039 | 4.55e-03 |
| GO:0032196 | 4.55e-03 |
| GO:0006313 | 4.55e-03 |
| GO:0007389 | 5.64e-03 |
| GO:0005575 | 2.20e-03 |
| GO:0032044 | 9.08e-03 |
| GO:0005737 | 1.30e-02 |
| GO:0071986 | 2.25e-02 |
| GO:0031231 | 2.25e-02 |
| GO:0005779 | 2.25e-02 |
| GO:0048500 | 2.70e-02 |
| GO:0003676 | 6.71e-05 |
| GO:1901363 | 3.39e-04 |
| GO:0097159 | 3.74e-04 |
| GO:0043167 | 3.46e-03 |
| GO:0046872 | 4.65e-03 |
| GO:0043169 | 5.13e-03 |
| GO:0008270 | 7.85e-03 |
| GO:0004958 | 9.08e-03 |
| GO:0050694 | 1.22e-02 |
| GO:0050698 | 1.22e-02 |
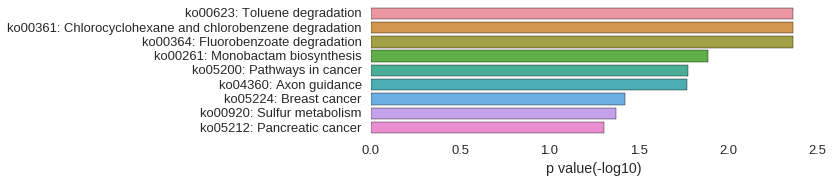
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12265
CAGCCCTCATGAACTTTCATTTTATATTTCTGTTTTAATTTCTTTTTTTCTTCCCCATAAAGACTCGAAC
AGAGCAGGACTCGTAATCAAGCGCCGCGCAGTCGTCCTGTGGATTTGCCAGGAATTATTCACCCATTATT
CATCCTCGTCCTTTATTTGTCCAAATCCTGGAAGAAGAAGAGGAAAAGAAATCAGCGCTTTGACCTCCAG
CATTAACAGAAGAGAAGAGCTTTGGTTTAAGCAGCAGCAGCAACAGCTCGCTATTCGGGATTTTATCACT
TTTGAGGATCGAGTTGGTTCACCTTCTTCACCTGAGTGAAACGGTAAATGGCTCTGTG
CAGCCCTCATGAACTTTCATTTTATATTTCTGTTTTAATTTCTTTTTTTCTTCCCCATAAAGACTCGAAC
AGAGCAGGACTCGTAATCAAGCGCCGCGCAGTCGTCCTGTGGATTTGCCAGGAATTATTCACCCATTATT
CATCCTCGTCCTTTATTTGTCCAAATCCTGGAAGAAGAAGAGGAAAAGAAATCAGCGCTTTGACCTCCAG
CATTAACAGAAGAGAAGAGCTTTGGTTTAAGCAGCAGCAGCAACAGCTCGCTATTCGGGATTTTATCACT
TTTGAGGATCGAGTTGGTTCACCTTCTTCACCTGAGTGAAACGGTAAATGGCTCTGTG