ZFLNCG12373
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1104059 | heart | kctd10 mut | 4.10 |
| SRR1004786 | larvae | control morpholino | 3.15 |
| SRR1104058 | heart | normal | 2.62 |
| SRR1004785 | larvae | normal | 2.31 |
| SRR610740 | 3 dpf | normal | 1.87 |
| SRR038627 | embryo | control morpholino | 1.54 |
| SRR726540 | 5 dpf | normal | 1.47 |
| SRR1004787 | larvae | impdh1a morpholino | 1.34 |
| SRR610741 | 3 dpf | express Ras | 1.31 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 1.17 |
Express in tissues
Correlated coding gene
Download
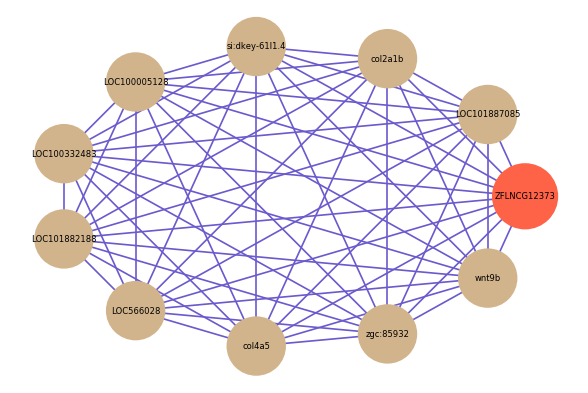
| gene | correlation coefficent |
|---|---|
| LOC101887085 | 0.65 |
| col2a1b | 0.63 |
| si:dkey-61l1.4 | 0.63 |
| LOC100005128 | 0.62 |
| LOC100332483 | 0.62 |
| LOC101882188 | 0.61 |
| LOC566028 | 0.61 |
| col4a5 | 0.61 |
| zgc:85932 | 0.61 |
| wnt9b | 0.61 |
Gene Ontology
Download
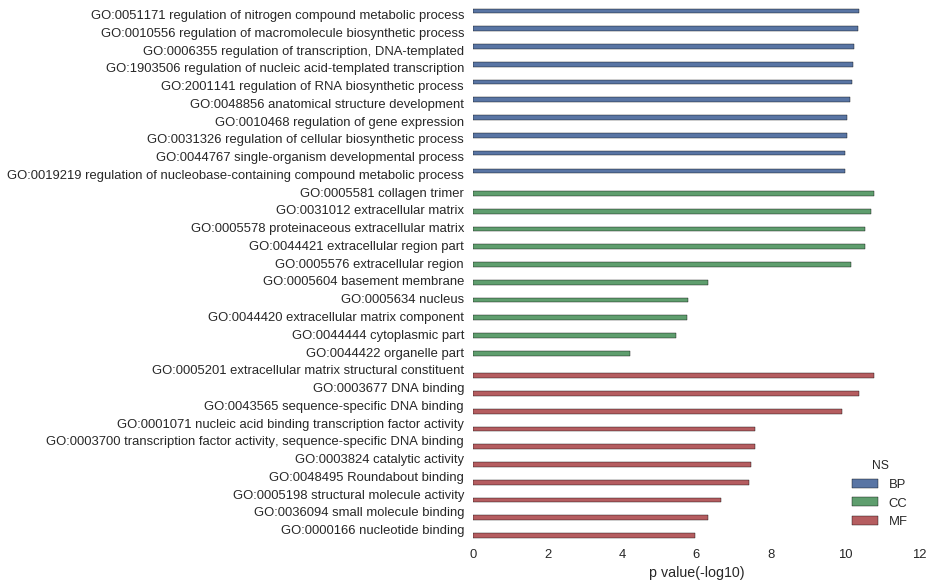
| GO | P value |
|---|---|
| GO:0051171 | 4.21e-11 |
| GO:0010556 | 4.42e-11 |
| GO:0006355 | 5.92e-11 |
| GO:1903506 | 6.06e-11 |
| GO:2001141 | 6.36e-11 |
| GO:0048856 | 7.42e-11 |
| GO:0010468 | 8.73e-11 |
| GO:0031326 | 9.13e-11 |
| GO:0044767 | 9.85e-11 |
| GO:0019219 | 1.00e-10 |
| GO:0005581 | 1.71e-11 |
| GO:0031012 | 2.04e-11 |
| GO:0005578 | 2.89e-11 |
| GO:0044421 | 2.91e-11 |
| GO:0005576 | 7.13e-11 |
| GO:0005604 | 4.96e-07 |
| GO:0005634 | 1.71e-06 |
| GO:0044420 | 1.75e-06 |
| GO:0044444 | 3.63e-06 |
| GO:0044422 | 5.91e-05 |
| GO:0005201 | 1.66e-11 |
| GO:0003677 | 4.23e-11 |
| GO:0043565 | 1.23e-10 |
| GO:0001071 | 2.60e-08 |
| GO:0003700 | 2.60e-08 |
| GO:0003824 | 3.40e-08 |
| GO:0048495 | 3.78e-08 |
| GO:0005198 | 2.19e-07 |
| GO:0036094 | 4.81e-07 |
| GO:0000166 | 1.10e-06 |
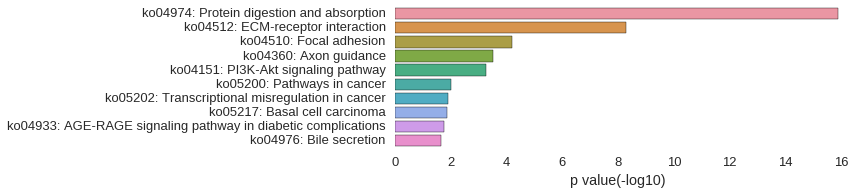
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12373
TGCAGACGAGAATTAAGTTTGTTGTATTTGTTCATCAGAAACAAACACTTGGATATTTTGGTAAGATGCA
TTATCACACACAAACATTTTCATATGATATATTTTTGTTGTTTTCTTTTTTCCTTATCTCATACATGTTA
TGGAAATTACATATATTTCTGTATGTACGTGTTGTATTTTAGAGAGCACACTGGTGAAGACTCAAGAGAG
GATGGGGAAACCTGTCCTGTTGTATGTTATAGGACTGCTGTCTTTTCTTGCCAGTAAGCCAAAATTTCAA
TATTAAATATTACAATAATCATAACTCACTTGTCCATTATTAATATTTAGATATTAAGTGATAAGCTGTT
TTTATGATGTAAAACAGAACATAACTCTTCTGTTCTTTCTCCATTAGGTGAAACAGATGCAACAG
TGCAGACGAGAATTAAGTTTGTTGTATTTGTTCATCAGAAACAAACACTTGGATATTTTGGTAAGATGCA
TTATCACACACAAACATTTTCATATGATATATTTTTGTTGTTTTCTTTTTTCCTTATCTCATACATGTTA
TGGAAATTACATATATTTCTGTATGTACGTGTTGTATTTTAGAGAGCACACTGGTGAAGACTCAAGAGAG
GATGGGGAAACCTGTCCTGTTGTATGTTATAGGACTGCTGTCTTTTCTTGCCAGTAAGCCAAAATTTCAA
TATTAAATATTACAATAATCATAACTCACTTGTCCATTATTAATATTTAGATATTAAGTGATAAGCTGTT
TTTATGATGTAAAACAGAACATAACTCTTCTGTTCTTTCTCCATTAGGTGAAACAGATGCAACAG