ZFLNCG12421
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 277.06 |
| SRR891495 | heart | normal | 66.13 |
| SRR749515 | 24 hpf | eif3ha morphant | 24.96 |
| SRR957181 | heart | normal | 24.05 |
| SRR749514 | 24 hpf | normal | 20.82 |
| SRR527836 | 24hpf | normal | 20.82 |
| SRR527844 | 24hpf | normal | 18.82 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 17.85 |
| SRR038627 | embryo | control morpholino | 15.87 |
| SRR700534 | heart | control morpholino | 15.73 |
Express in tissues
Correlated coding gene
Download
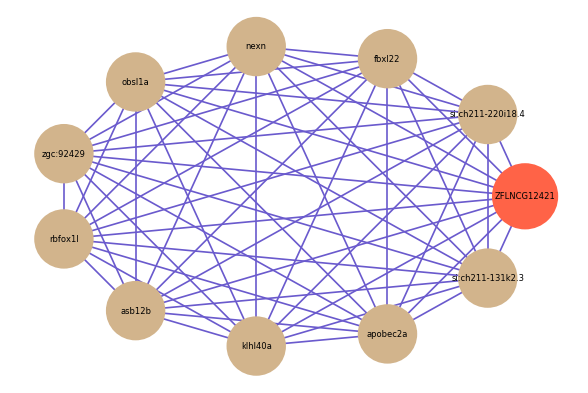
| gene | correlation coefficent |
|---|---|
| si:ch211-220i18.4 | 0.67 |
| fbxl22 | 0.66 |
| nexn | 0.65 |
| obsl1a | 0.65 |
| zgc:92429 | 0.65 |
| rbfox1l | 0.65 |
| asb12b | 0.64 |
| klhl40a | 0.63 |
| apobec2a | 0.62 |
| si:ch211-131k2.3 | 0.62 |
Gene Ontology
Download
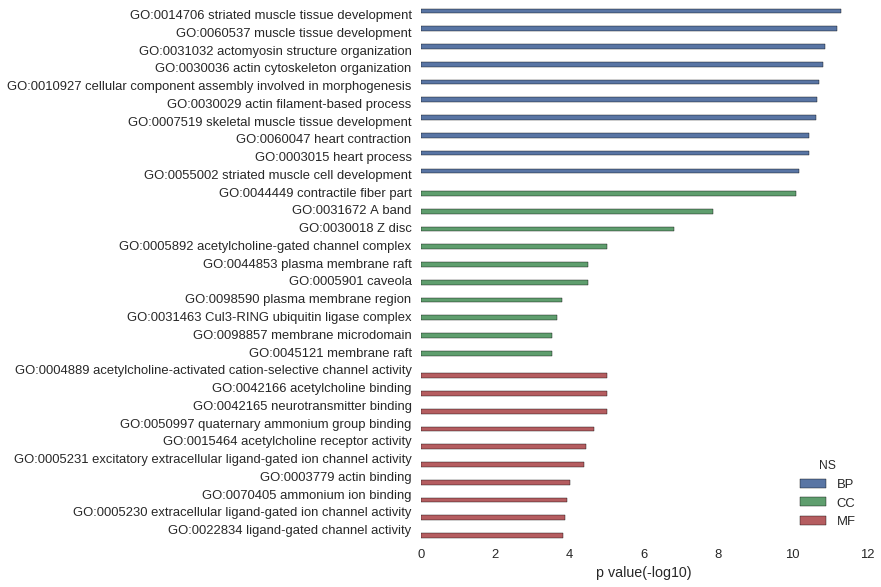
| GO | P value |
|---|---|
| GO:0014706 | 5.18e-12 |
| GO:0060537 | 6.29e-12 |
| GO:0031032 | 1.32e-11 |
| GO:0030036 | 1.52e-11 |
| GO:0010927 | 2.01e-11 |
| GO:0030029 | 2.21e-11 |
| GO:0007519 | 2.42e-11 |
| GO:0060047 | 3.72e-11 |
| GO:0003015 | 3.72e-11 |
| GO:0055002 | 6.72e-11 |
| GO:0044449 | 8.26e-11 |
| GO:0031672 | 1.35e-08 |
| GO:0030018 | 1.51e-07 |
| GO:0005892 | 9.64e-06 |
| GO:0044853 | 3.10e-05 |
| GO:0005901 | 3.10e-05 |
| GO:0098590 | 1.58e-04 |
| GO:0031463 | 2.23e-04 |
| GO:0098857 | 2.87e-04 |
| GO:0045121 | 2.87e-04 |
| GO:0004889 | 9.64e-06 |
| GO:0042166 | 9.64e-06 |
| GO:0042165 | 9.64e-06 |
| GO:0050997 | 2.15e-05 |
| GO:0015464 | 3.58e-05 |
| GO:0005231 | 4.14e-05 |
| GO:0003779 | 9.77e-05 |
| GO:0070405 | 1.19e-04 |
| GO:0005230 | 1.28e-04 |
| GO:0022834 | 1.46e-04 |
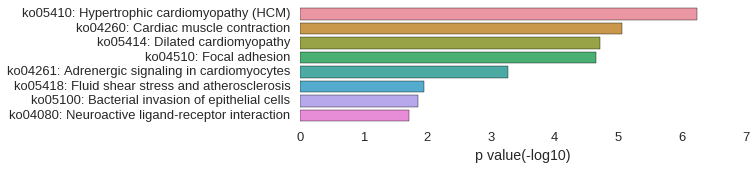
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12421
TTAATCCTGACCTCTTCTCTGGAAATCTTTATCACTTGATGAATGAGAACACAGTATAATGATGATCACA
GTGGTTTATTTATATGAAATACGTGAGTGCGAGTCCATAGTGTGACGCAGAGCACATAAAAGCAGCAGCA
TCAGTTGAGTAATAAGATAATCAGTCGGTATCAAGTAAGACACAAACCCCCTTTTGTTATCATGTTGTAA
TAATCTGTTGCTTTACTTTTATTTTAT
TTAATCCTGACCTCTTCTCTGGAAATCTTTATCACTTGATGAATGAGAACACAGTATAATGATGATCACA
GTGGTTTATTTATATGAAATACGTGAGTGCGAGTCCATAGTGTGACGCAGAGCACATAAAAGCAGCAGCA
TCAGTTGAGTAATAAGATAATCAGTCGGTATCAAGTAAGACACAAACCCCCTTTTGTTATCATGTTGTAA
TAATCTGTTGCTTTACTTTTATTTTAT