ZFLNCG12636
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941753 | posterior pectoral fin | normal | 12.20 |
| SRR038627 | embryo | control morpholino | 9.48 |
| SRR749515 | 24 hpf | eif3ha morphant | 9.32 |
| SRR941749 | anterior pectoral fin | normal | 7.45 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 6.95 |
| SRR1028004 | head | normal | 6.45 |
| SRR527832 | 16-36 hpf | normal | 6.39 |
| SRR749514 | 24 hpf | normal | 6.19 |
| SRR726542 | 5 dpf | infection with control | 6.13 |
| SRR516129 | skin | male and 5 month | 5.94 |
Express in tissues
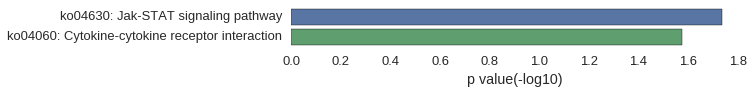
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| LOC101885379 | 0.58 |
| napgl | 0.53 |
| nr5a1a | 0.52 |
| v-fbpl | 0.52 |
| LOC101884213 | 0.52 |
| crfb9 | 0.51 |
| cldn10b | 0.50 |
Gene Ontology
Download
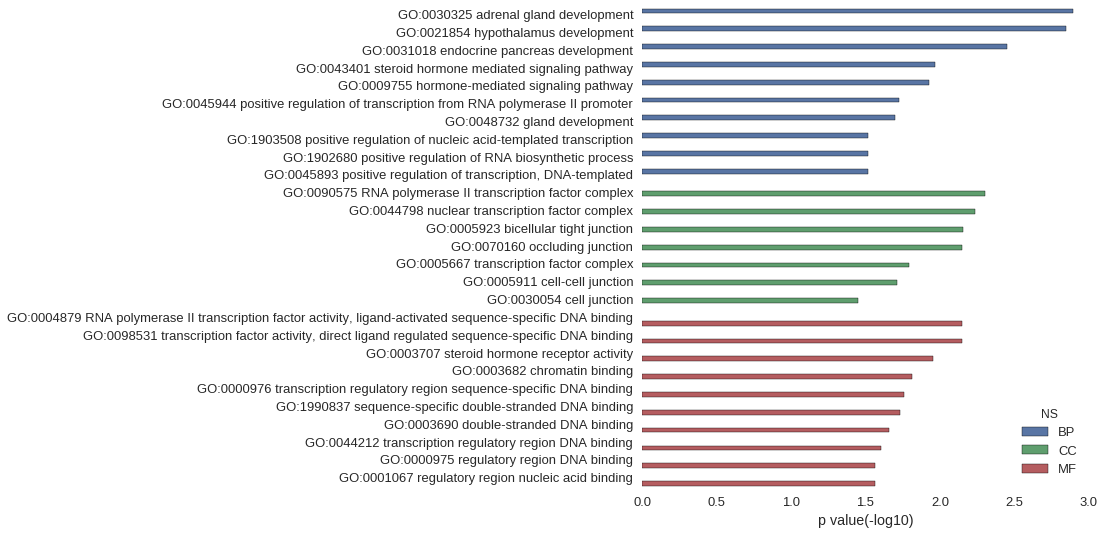
| GO | P value |
|---|---|
| GO:0030325 | 1.28e-03 |
| GO:0021854 | 1.42e-03 |
| GO:0031018 | 3.55e-03 |
| GO:0043401 | 1.08e-02 |
| GO:0009755 | 1.18e-02 |
| GO:0045944 | 1.88e-02 |
| GO:0048732 | 1.99e-02 |
| GO:1903508 | 3.05e-02 |
| GO:1902680 | 3.05e-02 |
| GO:0045893 | 3.05e-02 |
| GO:0090575 | 4.97e-03 |
| GO:0044798 | 5.82e-03 |
| GO:0005923 | 6.95e-03 |
| GO:0070160 | 7.09e-03 |
| GO:0005667 | 1.61e-02 |
| GO:0005911 | 1.94e-02 |
| GO:0030054 | 3.56e-02 |
| GO:0004879 | 7.09e-03 |
| GO:0098531 | 7.09e-03 |
| GO:0003707 | 1.11e-02 |
| GO:0003682 | 1.53e-02 |
| GO:0000976 | 1.75e-02 |
| GO:1990837 | 1.84e-02 |
| GO:0003690 | 2.19e-02 |
| GO:0044212 | 2.49e-02 |
| GO:0000975 | 2.71e-02 |
| GO:0001067 | 2.71e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12636
ACAGGAAACTAAAAGAAGTTGACAATAAACGCCTTTTTTGCTCTCAATCATGTACTAAACTCCTTGACAT
GTAATCTGTCAATAGTTTGTAGCAGAGATAATGCCGGCTGCCACGAGTGAGGTTGATTGGCAGCTTGGCT
TGGCCCCTCCCTCCTGTGGTTTGACGGGGGGTAGGTGAGAGAATACAGAGCCGGCCCCTTCCTCGCCCGC
TCCTCCCTGCCTCTCTCTCTCTCTATCTCTATCTCACTCTCTCTCTTGCTCTTGGTCTACGGTTCTGCTC
CTGTACACACGCACACACACCAAGC
ACAGGAAACTAAAAGAAGTTGACAATAAACGCCTTTTTTGCTCTCAATCATGTACTAAACTCCTTGACAT
GTAATCTGTCAATAGTTTGTAGCAGAGATAATGCCGGCTGCCACGAGTGAGGTTGATTGGCAGCTTGGCT
TGGCCCCTCCCTCCTGTGGTTTGACGGGGGGTAGGTGAGAGAATACAGAGCCGGCCCCTTCCTCGCCCGC
TCCTCCCTGCCTCTCTCTCTCTCTATCTCTATCTCACTCTCTCTCTTGCTCTTGGTCTACGGTTCTGCTC
CTGTACACACGCACACACACCAAGC