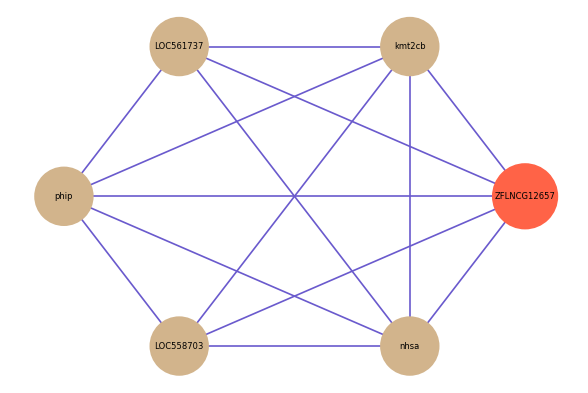
ZFLNCG12657
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR594771 | endothelium | normal | 4.71 |
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 1.23 |
| SRR514028 | retina | control morpholino | 1.02 |
| SRR800045 | muscle | normal | 0.85 |
| SRR1569491 | embryo | normal | 0.72 |
| SRR594769 | blood | normal | 0.67 |
| SRR514030 | retina | id2a morpholino | 0.65 |
| SRR592699 | pineal gland | normal | 0.54 |
| SRR700534 | heart | control morpholino | 0.53 |
| ERR023144 | brain | normal | 0.41 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
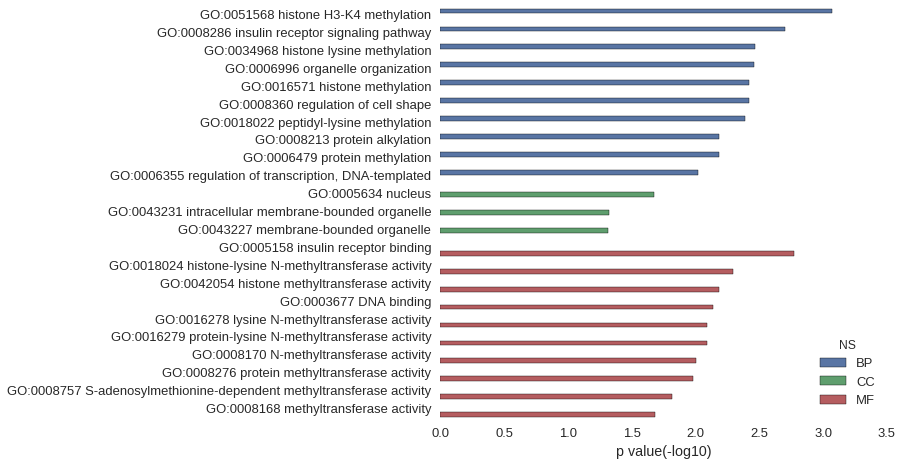
| GO | P value |
|---|---|
| GO:0051568 | 8.53e-04 |
| GO:0008286 | 1.99e-03 |
| GO:0034968 | 3.41e-03 |
| GO:0006996 | 3.51e-03 |
| GO:0016571 | 3.83e-03 |
| GO:0008360 | 3.83e-03 |
| GO:0018022 | 4.12e-03 |
| GO:0008213 | 6.53e-03 |
| GO:0006479 | 6.53e-03 |
| GO:0006355 | 9.64e-03 |
| GO:0005634 | 2.13e-02 |
| GO:0043231 | 4.79e-02 |
| GO:0043227 | 4.86e-02 |
| GO:0005158 | 1.70e-03 |
| GO:0018024 | 5.11e-03 |
| GO:0042054 | 6.53e-03 |
| GO:0003677 | 7.33e-03 |
| GO:0016278 | 8.09e-03 |
| GO:0016279 | 8.09e-03 |
| GO:0008170 | 9.92e-03 |
| GO:0008276 | 1.05e-02 |
| GO:0008757 | 1.53e-02 |
| GO:0008168 | 2.09e-02 |
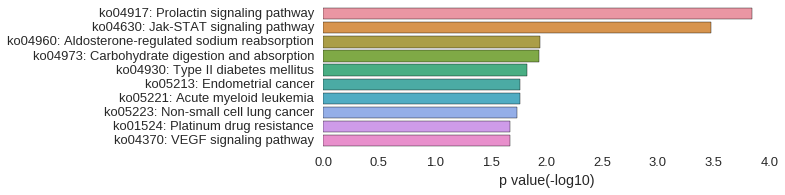
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12657
AGAAAGTTCACCGCATGACTCCATTTCATGGACAATTTTTAAGGGTTATTTCAGGTCAAATTGAGTTTAG
ACACATACCTTTTTCACAATTCCTGTTTAATTCTCAAAAAGTTTTGGTCATTAAAGCAAAACTGCAGCTT
TTCTTAGTATAACATGCATAAAACCAAGAAAAACACAAATACAACATTACAAAAAATTATATTTGCAAAG
ATCTTTTCCATTCTCCCACACAGTCATTTGTATCACTGACCCATTTAAACCCATATAAATAAACAGTTAA
GCAGAAACACTCAATTTGGGCACAAACCTATCATCCAGTTCCTCTGTCTTGCGTTATCAAACTGCTGCCG
AAGAACCAGGAAGTCGATGACATCAGGCATGTCGTGATATCTGAGTCACCAATAGAGATGCAAGGCAAGG
GTTAAAAGCCACTCAGCACAGATCATGTTGAATAAGAGCCTGTGACTCACGCAGACTGAACTCACTTAAT
GGAGAATGAGCCGCCCGTCAGTTTGCCTGAGTCTGGGTCCAGGAAGGCCAGCTTCAGACAGCACAGAGTA
GGCAAACCCACCTCGTACTTAATGCCCACAATCTTCAACAGCTCCTGCTCCTATAGGCACACAATCACAA
AGAAAGTTCACCGCATGACTCCATTTCATGGACAATTTTTAAGGGTTATTTCAGGTCAAATTGAGTTTAG
ACACATACCTTTTTCACAATTCCTGTTTAATTCTCAAAAAGTTTTGGTCATTAAAGCAAAACTGCAGCTT
TTCTTAGTATAACATGCATAAAACCAAGAAAAACACAAATACAACATTACAAAAAATTATATTTGCAAAG
ATCTTTTCCATTCTCCCACACAGTCATTTGTATCACTGACCCATTTAAACCCATATAAATAAACAGTTAA
GCAGAAACACTCAATTTGGGCACAAACCTATCATCCAGTTCCTCTGTCTTGCGTTATCAAACTGCTGCCG
AAGAACCAGGAAGTCGATGACATCAGGCATGTCGTGATATCTGAGTCACCAATAGAGATGCAAGGCAAGG
GTTAAAAGCCACTCAGCACAGATCATGTTGAATAAGAGCCTGTGACTCACGCAGACTGAACTCACTTAAT
GGAGAATGAGCCGCCCGTCAGTTTGCCTGAGTCTGGGTCCAGGAAGGCCAGCTTCAGACAGCACAGAGTA
GGCAAACCCACCTCGTACTTAATGCCCACAATCTTCAACAGCTCCTGCTCCTATAGGCACACAATCACAA