ZFLNCG12757
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516124 | skin | male and 3.5 year | 25.09 |
| SRR941753 | posterior pectoral fin | normal | 23.81 |
| SRR516129 | skin | male and 5 month | 23.14 |
| SRR516123 | skin | male and 3.5 year | 19.39 |
| SRR941749 | anterior pectoral fin | normal | 16.78 |
| SRR516125 | skin | male and 3.5 year | 14.88 |
| SRR592702 | pineal gland | normal | 14.65 |
| SRR1299124 | caudal fin | Zero day time after treatment | 14.42 |
| SRR891512 | blood | normal | 8.96 |
| SRR942767 | embryo | myd88-/- and infection with Mycobacterium marinum | 8.84 |
Express in tissues
Correlated coding gene
Download
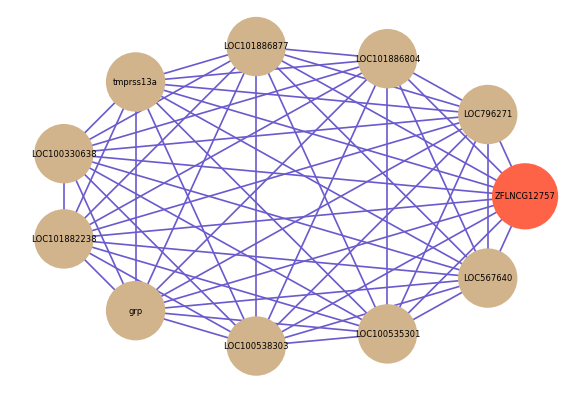
| gene | correlation coefficent |
|---|---|
| LOC796271 | 0.62 |
| LOC101886804 | 0.62 |
| LOC101886877 | 0.61 |
| tmprss13a | 0.60 |
| LOC100330638 | 0.59 |
| LOC101882238 | 0.59 |
| grp | 0.59 |
| LOC100538303 | 0.58 |
| LOC100535301 | 0.58 |
| LOC567640 | 0.58 |
Gene Ontology
Download
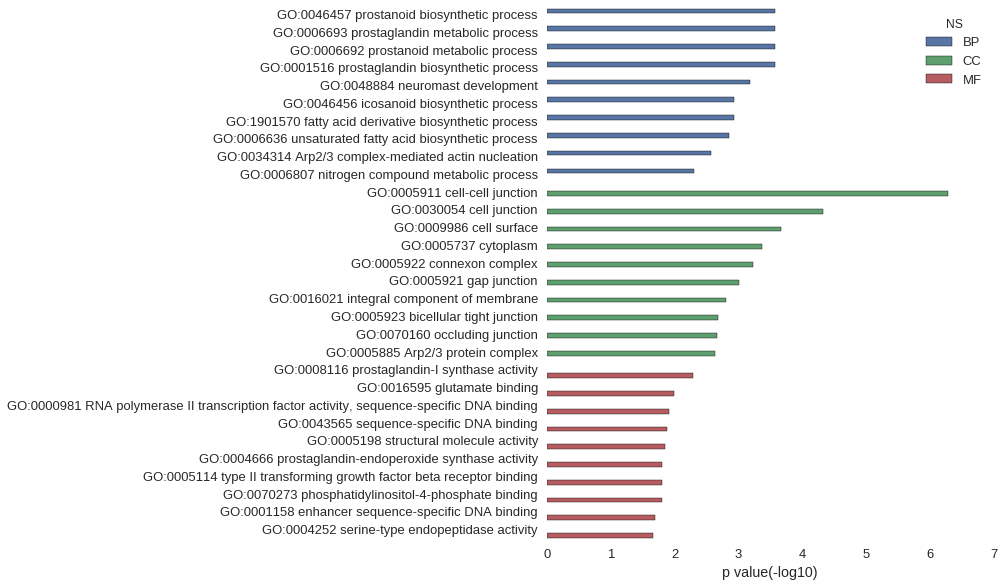
| GO | P value |
|---|---|
| GO:0046457 | 2.63e-04 |
| GO:0006693 | 2.63e-04 |
| GO:0006692 | 2.63e-04 |
| GO:0001516 | 2.63e-04 |
| GO:0048884 | 6.53e-04 |
| GO:0046456 | 1.16e-03 |
| GO:1901570 | 1.16e-03 |
| GO:0006636 | 1.42e-03 |
| GO:0034314 | 2.67e-03 |
| GO:0006807 | 4.94e-03 |
| GO:0005911 | 5.19e-07 |
| GO:0030054 | 4.79e-05 |
| GO:0009986 | 2.14e-04 |
| GO:0005737 | 4.31e-04 |
| GO:0005922 | 5.96e-04 |
| GO:0005921 | 9.92e-04 |
| GO:0016021 | 1.58e-03 |
| GO:0005923 | 2.08e-03 |
| GO:0070160 | 2.20e-03 |
| GO:0005885 | 2.32e-03 |
| GO:0008116 | 5.19e-03 |
| GO:0016595 | 1.03e-02 |
| GO:0000981 | 1.21e-02 |
| GO:0043565 | 1.33e-02 |
| GO:0005198 | 1.39e-02 |
| GO:0004666 | 1.55e-02 |
| GO:0005114 | 1.55e-02 |
| GO:0070273 | 1.55e-02 |
| GO:0001158 | 2.06e-02 |
| GO:0004252 | 2.19e-02 |
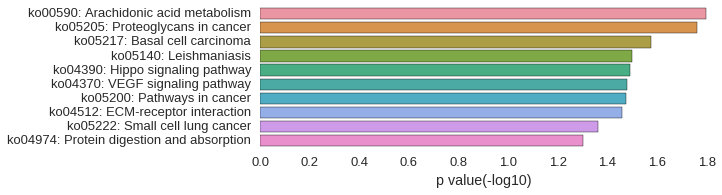
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12757
GATATTAAAGTGGTGTTTACACCATTTCTGCATTTTGAAATCTCGGCAACTGGAGCTGGAGCTGGAGGTT
TGTAGTACTCAGCATGTTGTTGCAGTGTTTACAGTGCTGATCCTATACTTCTGGGTTTCTTCCCACCCAG
CCTTGCTTTCGTGTTTTAAAATGGCGGGCTCATGAAATAAGCGGCGGATGTATGATAGGGCCCAGAGTAT
GTGTGAAACAGGCGCA
GATATTAAAGTGGTGTTTACACCATTTCTGCATTTTGAAATCTCGGCAACTGGAGCTGGAGCTGGAGGTT
TGTAGTACTCAGCATGTTGTTGCAGTGTTTACAGTGCTGATCCTATACTTCTGGGTTTCTTCCCACCCAG
CCTTGCTTTCGTGTTTTAAAATGGCGGGCTCATGAAATAAGCGGCGGATGTATGATAGGGCCCAGAGTAT
GTGTGAAACAGGCGCA