ZFLNCG12768
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372791 | dome | normal | 28.68 |
| SRR372787 | 2-4 cell | normal | 24.54 |
| SRR372793 | shield | normal | 21.18 |
| ERR023144 | brain | normal | 20.94 |
| SRR1021215 | sphere stage | Mzeomesa | 18.65 |
| SRR1049814 | 256 cell | normal | 18.53 |
| SRR748489 | egg | normal | 17.19 |
| SRR1621206 | embryo | normal | 16.78 |
| SRR372789 | 1K cell | normal | 13.66 |
| SRR748491 | germ ring | normal | 11.64 |
Express in tissues
Correlated coding gene
Download
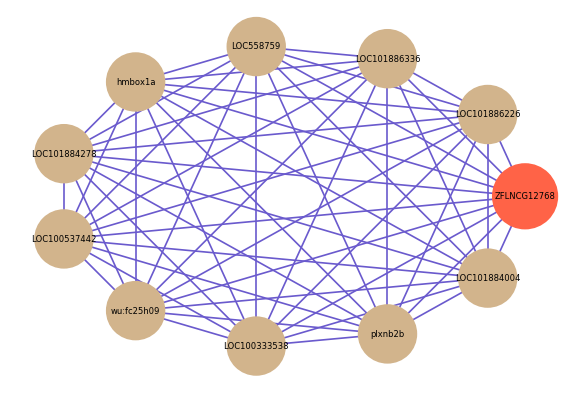
| gene | correlation coefficent |
|---|---|
| LOC101886226 | 0.70 |
| LOC101886336 | 0.66 |
| LOC558759 | 0.65 |
| hmbox1a | 0.64 |
| LOC101884278 | 0.64 |
| LOC100537442 | 0.64 |
| wu:fc25h09 | 0.63 |
| LOC100333538 | 0.63 |
| plxnb2b | 0.62 |
| LOC101884004 | 0.62 |
Gene Ontology
Download
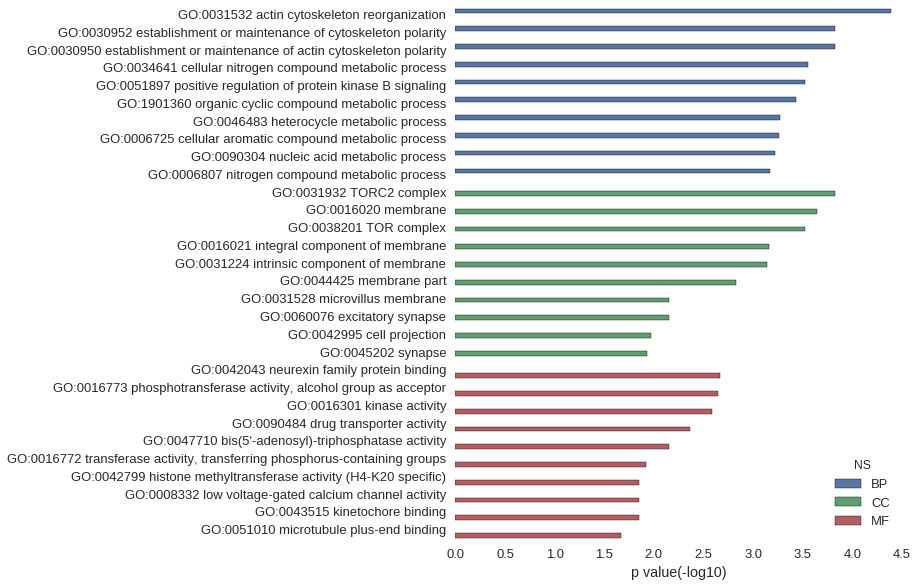
| GO | P value |
|---|---|
| GO:0031532 | 4.03e-05 |
| GO:0030952 | 1.49e-04 |
| GO:0030950 | 1.49e-04 |
| GO:0034641 | 2.78e-04 |
| GO:0051897 | 2.97e-04 |
| GO:1901360 | 3.70e-04 |
| GO:0046483 | 5.39e-04 |
| GO:0006725 | 5.44e-04 |
| GO:0090304 | 6.01e-04 |
| GO:0006807 | 6.74e-04 |
| GO:0031932 | 1.49e-04 |
| GO:0016020 | 2.28e-04 |
| GO:0038201 | 2.97e-04 |
| GO:0016021 | 6.95e-04 |
| GO:0031224 | 7.26e-04 |
| GO:0044425 | 1.47e-03 |
| GO:0031528 | 7.11e-03 |
| GO:0060076 | 7.11e-03 |
| GO:0042995 | 1.07e-02 |
| GO:0045202 | 1.18e-02 |
| GO:0042043 | 2.17e-03 |
| GO:0016773 | 2.24e-03 |
| GO:0016301 | 2.59e-03 |
| GO:0090484 | 4.30e-03 |
| GO:0047710 | 7.11e-03 |
| GO:0016772 | 1.19e-02 |
| GO:0042799 | 1.42e-02 |
| GO:0008332 | 1.42e-02 |
| GO:0043515 | 1.42e-02 |
| GO:0051010 | 2.12e-02 |
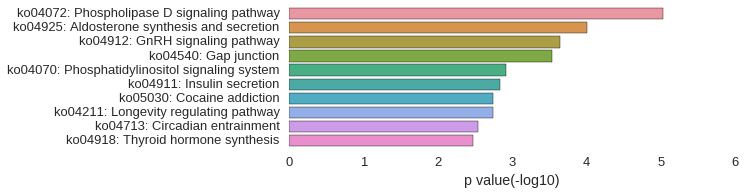
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12768
TATCTTTCATCTGGAAGGTGACGGCCATCACTTTGATCAGCTGTTTGCCGCTCTACATCCTCAAATACCT
GCGCAGACGCTTTTCTCCTCCCAGTTACTCCAAACTGACCTCTTAAACCCTTCACAAAAGAACAAAAAAC
CCTAAAACTACTGGAGCTCCAGACCTCCGTTCTCCATCATACTAAAGTAACAGGTCAGTTTATGTGGTAA
GGCAGGCAGTTTTTTTTTTCGTCTGAAATGAGCAGGTGAACATTAGCTTGAAGGCTGGACGCTTTTCTGT
TGGAGTCCAGACGCTGTTAGCTATGAATCATGTCATTTCTGAAGTGAGTTTAAGTTTGGGTGAATCACTG
GCCCCAAGGAAGACTAAACACACTGGCTAACTTTAAGGGGTGGACAGGAGACAACAG
TATCTTTCATCTGGAAGGTGACGGCCATCACTTTGATCAGCTGTTTGCCGCTCTACATCCTCAAATACCT
GCGCAGACGCTTTTCTCCTCCCAGTTACTCCAAACTGACCTCTTAAACCCTTCACAAAAGAACAAAAAAC
CCTAAAACTACTGGAGCTCCAGACCTCCGTTCTCCATCATACTAAAGTAACAGGTCAGTTTATGTGGTAA
GGCAGGCAGTTTTTTTTTTCGTCTGAAATGAGCAGGTGAACATTAGCTTGAAGGCTGGACGCTTTTCTGT
TGGAGTCCAGACGCTGTTAGCTATGAATCATGTCATTTCTGAAGTGAGTTTAAGTTTGGGTGAATCACTG
GCCCCAAGGAAGACTAAACACACTGGCTAACTTTAAGGGGTGGACAGGAGACAACAG