ZFLNCG12933
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR957180 | heart | 7 days after heart tip amputation | 326.25 |
| SRR957181 | heart | normal | 136.97 |
| SRR1035239 | liver | transgenic mCherry control | 66.38 |
| SRR372796 | bud | normal | 49.71 |
| ERR023144 | brain | normal | 43.52 |
| SRR1035237 | liver | transgenic UHRF1 | 43.07 |
| SRR658535 | bud | Gata5 morphant | 42.77 |
| ERR023143 | swim bladder | normal | 40.57 |
| SRR658533 | bud | normal | 40.47 |
| SRR700534 | heart | control morpholino | 38.34 |
Express in tissues
Correlated coding gene
Download
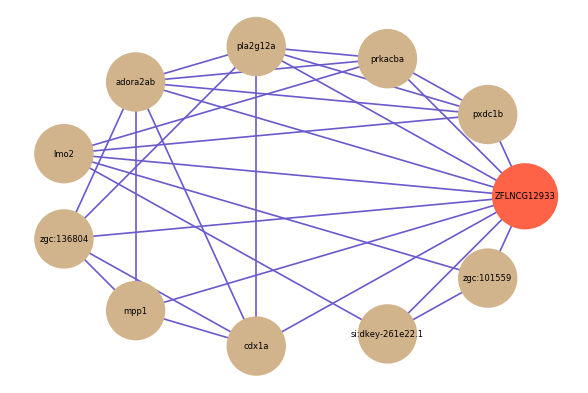
| gene | correlation coefficent |
|---|---|
| pxdc1b | 0.63 |
| zgc:136804 | 0.56 |
| prkacba | 0.55 |
| mpp1 | 0.54 |
| pla2g12a | 0.54 |
| si:dkey-261e22.1 | 0.54 |
| adora2ab | 0.54 |
| lmo2 | 0.53 |
| zgc:101559 | 0.53 |
| cdx1a | 0.51 |
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0061304 | 9.95e-04 |
| GO:0061299 | 9.95e-04 |
| GO:0070197 | 1.99e-03 |
| GO:0021906 | 1.99e-03 |
| GO:0097240 | 1.99e-03 |
| GO:0007129 | 1.99e-03 |
| GO:0017144 | 2.98e-03 |
| GO:0016559 | 2.98e-03 |
| GO:0044088 | 3.97e-03 |
| GO:2000785 | 3.97e-03 |
| GO:0031231 | 4.97e-03 |
| GO:0005779 | 4.97e-03 |
| GO:0000784 | 9.91e-03 |
| GO:0005637 | 1.29e-02 |
| GO:0044439 | 1.38e-02 |
| GO:0044438 | 1.38e-02 |
| GO:0000781 | 1.58e-02 |
| GO:0098687 | 3.81e-02 |
| GO:0031965 | 3.81e-02 |
| GO:0004499 | 3.97e-03 |
| GO:0001609 | 3.97e-03 |
| GO:0004623 | 4.97e-03 |
| GO:0003756 | 1.19e-02 |
| GO:0016864 | 1.19e-02 |
| GO:0035586 | 1.29e-02 |
| GO:0016709 | 1.58e-02 |
| GO:0050661 | 1.78e-02 |
| GO:0016860 | 3.04e-02 |
| GO:0050660 | 4.67e-02 |
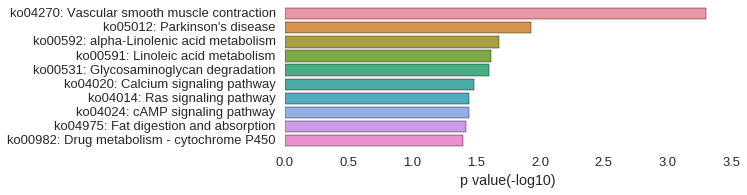
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12933
GGGGGCAAATAATTCTGACTTCAACTGTATATTTATGTAGGAATATACTAGGAGTCATGTGCAAAATGAG
TGAGACTCTCTCAAAACAGACAACCAACCCAACAGCAGACTTGACAACGCTCTGTTAACATGTTAAACAG
CTTGTGGTACAGGTCATAACTCCTGCTTTCTCCATCTAAATGAACAGAATGTGGAAATATAAATGTA
GGGGGCAAATAATTCTGACTTCAACTGTATATTTATGTAGGAATATACTAGGAGTCATGTGCAAAATGAG
TGAGACTCTCTCAAAACAGACAACCAACCCAACAGCAGACTTGACAACGCTCTGTTAACATGTTAAACAG
CTTGTGGTACAGGTCATAACTCCTGCTTTCTCCATCTAAATGAACAGAATGTGGAAATATAAATGTA