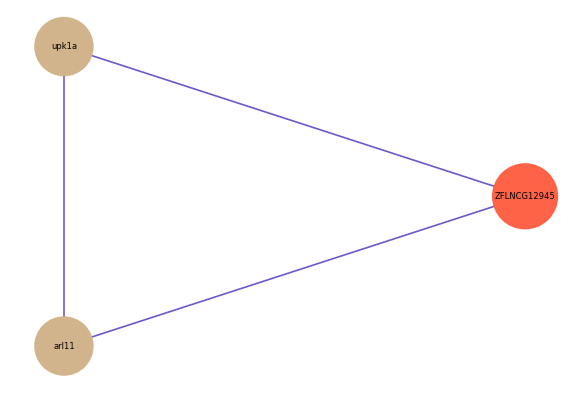
ZFLNCG12945
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1299126 | caudal fin | One day time after treatment | 180.76 |
| SRR1299127 | caudal fin | Two days time after treatment | 162.28 |
| SRR1299125 | caudal fin | Half day time after treatment | 159.24 |
| SRR1299129 | caudal fin | Seven days time after treatment | 103.44 |
| SRR1299128 | caudal fin | Three days time after treatment | 78.65 |
| SRR957180 | heart | 7 days after heart tip amputation | 67.32 |
| SRR957181 | heart | normal | 64.45 |
| SRR891512 | blood | normal | 57.62 |
| SRR516121 | skin | male and 3.5 year | 51.52 |
| SRR1299124 | caudal fin | Zero day time after treatment | 42.60 |
Express in tissues
Gene Ontology
Download
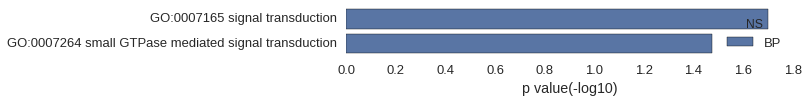
| GO | P value |
|---|---|
| GO:0007165 | 1.99e-02 |
| GO:0007264 | 3.35e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12945
AGGGTAAAAAACAGTTTTGTTGTTTCCTCAGGTACTACAGTGAGAGAGGAGACGCTGTAGCCAAGGCTTC
CAAACAGCCACATGTGGTCAGTTTAATTTATTGTTAGTTCTTTTTTTGTATATGCTTTGAGCTTCTGATA
ATGATTTTAACCCTGCTTTTCTTCTGGGCAACAGGGAGATTTCCGACAGCTTGTTCATGAACTGGATCAG
CACCAGTATTGTGAGTTACGCATCATAGTTCTGGAGATTCGCAACACTTACGTAAGTTCTGTCTCGGCTT
TCCCTGAAAACAACATCTCAGTGCTGCATTTCTCATCATTCATGTAAGTTGGTGAGTCTGTAAGATGGCT
GTGACTGATTTCATCCATTCCTTCGACTCTAGGCCATGCTGTATGATGTCATCACCAAGAACTTTGACAA
GATTAAGAAGCCCAGGGGAGACTTATCTTCAAAAGCACTTATCTACTGAGCAACCAGGCTGATGTGCACT
GAAATCACACACGCAAACACACACACATTTACTGAAAAGTAAAACATGCCGTATTGAGTTTCATAACCAC
AGTCACAGTGCAGTGACTTGAGATACGATTACATCAACAGAATGTGCTGTATTACAGTTTTTAATTCAAT
AAATAGTTTAGGCTA
AGGGTAAAAAACAGTTTTGTTGTTTCCTCAGGTACTACAGTGAGAGAGGAGACGCTGTAGCCAAGGCTTC
CAAACAGCCACATGTGGTCAGTTTAATTTATTGTTAGTTCTTTTTTTGTATATGCTTTGAGCTTCTGATA
ATGATTTTAACCCTGCTTTTCTTCTGGGCAACAGGGAGATTTCCGACAGCTTGTTCATGAACTGGATCAG
CACCAGTATTGTGAGTTACGCATCATAGTTCTGGAGATTCGCAACACTTACGTAAGTTCTGTCTCGGCTT
TCCCTGAAAACAACATCTCAGTGCTGCATTTCTCATCATTCATGTAAGTTGGTGAGTCTGTAAGATGGCT
GTGACTGATTTCATCCATTCCTTCGACTCTAGGCCATGCTGTATGATGTCATCACCAAGAACTTTGACAA
GATTAAGAAGCCCAGGGGAGACTTATCTTCAAAAGCACTTATCTACTGAGCAACCAGGCTGATGTGCACT
GAAATCACACACGCAAACACACACACATTTACTGAAAAGTAAAACATGCCGTATTGAGTTTCATAACCAC
AGTCACAGTGCAGTGACTTGAGATACGATTACATCAACAGAATGTGCTGTATTACAGTTTTTAATTCAAT
AAATAGTTTAGGCTA