ZFLNCG13081
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700537 | heart | Gata4 morpholino | 13.68 |
| SRR700534 | heart | control morpholino | 8.81 |
| SRR1647682 | spleen | normal | 4.79 |
| SRR516131 | skin | male and 5 month | 4.23 |
| SRR1647683 | spleen | SVCV treatment | 2.80 |
| SRR516122 | skin | male and 3.5 year | 2.35 |
| SRR1647684 | spleen | SVCV treatment | 1.94 |
| SRR516121 | skin | male and 3.5 year | 1.48 |
| SRR891512 | blood | normal | 1.23 |
| SRR800045 | muscle | normal | 1.18 |
Express in tissues
Correlated coding gene
Download
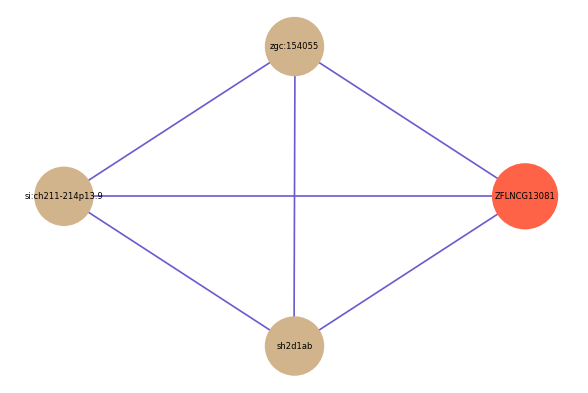
| gene | correlation coefficent |
|---|---|
| zgc:154055 | 0.54 |
| si:ch211-214p13.9 | 0.51 |
| sh2d1ab | 0.50 |
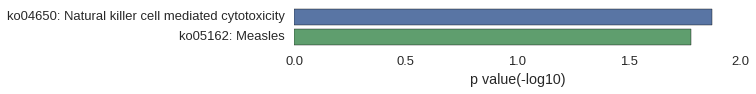
Gene Ontology
Download
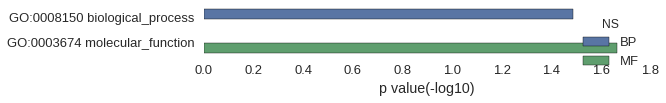
| GO | P value |
|---|---|
| GO:0008150 | 3.25e-02 |
| GO:0003674 | 2.16e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG13081
TTCCTTTAAAAAAAAAAATTTCAGCATAGAAAGAAGCAGGATTTTCCCAGTATGGGTTGCGACTGGAAGG
GCATCCGCTGCGTAAAACATATGCTGGATAGATTAGCGGTTTATTCTGCTGTAGTGACCCCAGATTAATA
AAGGGATTAATAAAGGGATTAATAATGGGATTAATAAAGGGACTAAGCCAAAAAGAAAAAGAATGAAAGA
ACCAGGATTGTATAGCGTAGCATAGGACCCCCCCCCCCTCTCTTCTTTGTAGTGTGGGTGCTGAAGTTCA
AACTCTGGATAACATTCACAGCTGCCTAGTTTTCAGTGTTTATTTTTACACTTTATCCCCTTCTGTTGAA
TAAGAACAACAAAAACAACAACATAAATCTGTTTAGAACGTGTTTTTGCTCGATTTGCCAATACGATTGC
AATATGTTTGTCTAAATCCTTGTGAGAGGTTTATGTATTATATTCAGTTTAGCTTTTCCATATTACATTT
TATTATATTTACAATTCTAACCTTTTCTAGATGTTCTATCTTTGCACTGAGCCAAGGTGCTTTAAGATTT
ATACAGCACATCCTGTTCTTTTTATCAA
TTCCTTTAAAAAAAAAAATTTCAGCATAGAAAGAAGCAGGATTTTCCCAGTATGGGTTGCGACTGGAAGG
GCATCCGCTGCGTAAAACATATGCTGGATAGATTAGCGGTTTATTCTGCTGTAGTGACCCCAGATTAATA
AAGGGATTAATAAAGGGATTAATAATGGGATTAATAAAGGGACTAAGCCAAAAAGAAAAAGAATGAAAGA
ACCAGGATTGTATAGCGTAGCATAGGACCCCCCCCCCCTCTCTTCTTTGTAGTGTGGGTGCTGAAGTTCA
AACTCTGGATAACATTCACAGCTGCCTAGTTTTCAGTGTTTATTTTTACACTTTATCCCCTTCTGTTGAA
TAAGAACAACAAAAACAACAACATAAATCTGTTTAGAACGTGTTTTTGCTCGATTTGCCAATACGATTGC
AATATGTTTGTCTAAATCCTTGTGAGAGGTTTATGTATTATATTCAGTTTAGCTTTTCCATATTACATTT
TATTATATTTACAATTCTAACCTTTTCTAGATGTTCTATCTTTGCACTGAGCCAAGGTGCTTTAAGATTT
ATACAGCACATCCTGTTCTTTTTATCAA