ZFLNCG13154
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR658533 | bud | normal | 61.93 |
| SRR658539 | bud | Gata5/6 morphant | 54.81 |
| SRR658537 | bud | Gata6 morphant | 46.28 |
| SRR658535 | bud | Gata5 morphant | 41.70 |
| SRR658545 | 6 somite | Gata6 morphant | 39.57 |
| SRR658547 | 6 somite | Gata5/6 morphant | 38.37 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 31.82 |
| SRR1035984 | 13 hpf | normal | 29.83 |
| SRR749515 | 24 hpf | eif3ha morphant | 29.76 |
| SRR1035978 | 13 hpf | rx3-/- | 28.16 |
Express in tissues
Correlated coding gene
Download
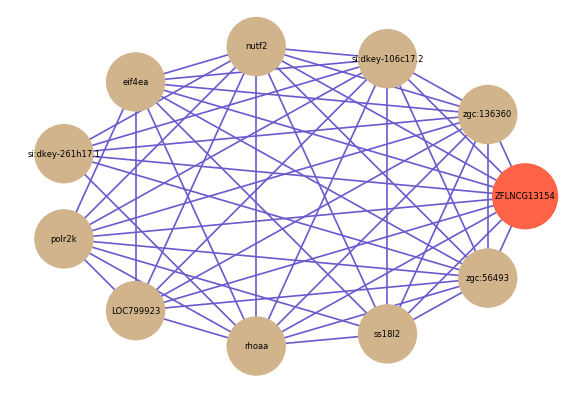
| gene | correlation coefficent |
|---|---|
| zgc:136360 | 0.61 |
| si:dkey-106c17.2 | 0.60 |
| nutf2 | 0.60 |
| eif4ea | 0.60 |
| si:dkey-261h17.1 | 0.60 |
| polr2k | 0.60 |
| LOC799923 | 0.59 |
| rhoaa | 0.59 |
| ss18l2 | 0.59 |
| zgc:56493 | 0.58 |
Gene Ontology
Download
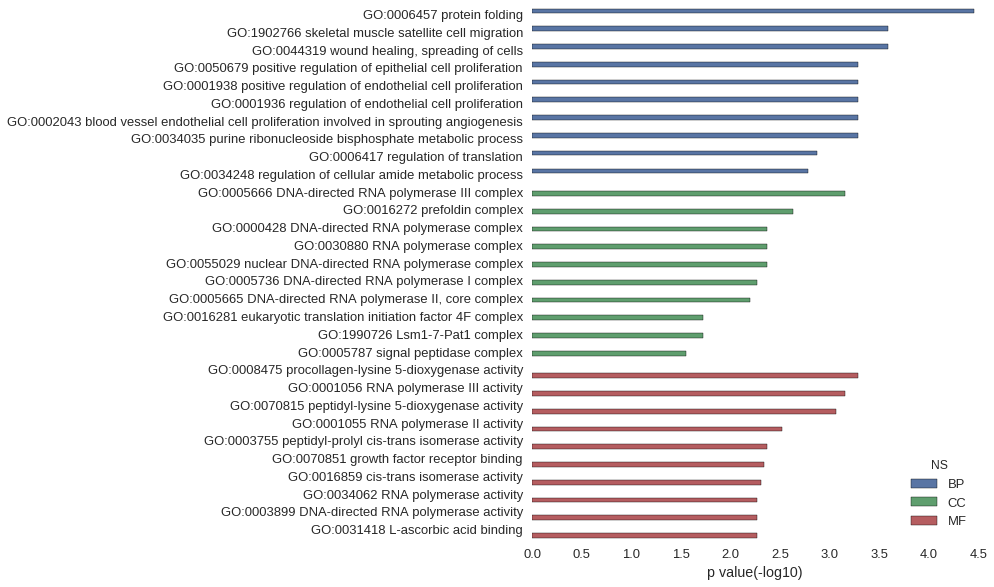
| GO | P value |
|---|---|
| GO:0006457 | 3.47e-05 |
| GO:1902766 | 2.60e-04 |
| GO:0044319 | 2.60e-04 |
| GO:0050679 | 5.18e-04 |
| GO:0001938 | 5.18e-04 |
| GO:0001936 | 5.18e-04 |
| GO:0002043 | 5.18e-04 |
| GO:0034035 | 5.18e-04 |
| GO:0006417 | 1.34e-03 |
| GO:0034248 | 1.64e-03 |
| GO:0005666 | 7.00e-04 |
| GO:0016272 | 2.36e-03 |
| GO:0000428 | 4.24e-03 |
| GO:0030880 | 4.24e-03 |
| GO:0055029 | 4.24e-03 |
| GO:0005736 | 5.42e-03 |
| GO:0005665 | 6.37e-03 |
| GO:0016281 | 1.87e-02 |
| GO:1990726 | 1.87e-02 |
| GO:0005787 | 2.79e-02 |
| GO:0008475 | 5.18e-04 |
| GO:0001056 | 7.00e-04 |
| GO:0070815 | 8.57e-04 |
| GO:0001055 | 3.01e-03 |
| GO:0003755 | 4.24e-03 |
| GO:0070851 | 4.59e-03 |
| GO:0016859 | 4.97e-03 |
| GO:0034062 | 5.36e-03 |
| GO:0003899 | 5.36e-03 |
| GO:0031418 | 5.42e-03 |
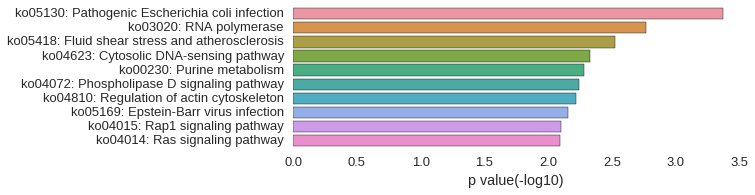
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG13154
GAAAAACAAAACTAAAAATACAGAAAACTGCCAGAACAGTACTCGAGGGGGACACACAAATGAGTCATTC
TCTGCAAGTTTATTTTATGTAAATGCTACTGCACGCTTTCGTCTCCTCAAGGAAAATGCAGAGTACTAAA
AGCATTGTTTAATCAGGCATACTACATCACACACATCATTTCAATCAGGCCTAAAAATTCAACACTCAAT
TGGTAGCTGCTTTGCTGACATTCTCAGTGGCACTGTGGTATTCAAAAACACATTCATAATAATGCAGCAA
TATCTGAC
GAAAAACAAAACTAAAAATACAGAAAACTGCCAGAACAGTACTCGAGGGGGACACACAAATGAGTCATTC
TCTGCAAGTTTATTTTATGTAAATGCTACTGCACGCTTTCGTCTCCTCAAGGAAAATGCAGAGTACTAAA
AGCATTGTTTAATCAGGCATACTACATCACACACATCATTTCAATCAGGCCTAAAAATTCAACACTCAAT
TGGTAGCTGCTTTGCTGACATTCTCAGTGGCACTGTGGTATTCAAAAACACATTCATAATAATGCAGCAA
TATCTGAC