ZFLNCG13235
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516134 | skin | male and 5 month | 114.75 |
| SRR516131 | skin | male and 5 month | 72.80 |
| SRR516133 | skin | male and 5 month | 70.66 |
| SRR516130 | skin | male and 5 month | 56.10 |
| SRR516126 | skin | male and 5 month | 49.00 |
| SRR516127 | skin | male and 5 month | 47.12 |
| SRR516129 | skin | male and 5 month | 41.94 |
| SRR516132 | skin | male and 5 month | 41.01 |
| SRR516135 | skin | male and 5 month | 20.73 |
| SRR516122 | skin | male and 3.5 year | 20.26 |
Express in tissues
Correlated coding gene
Download
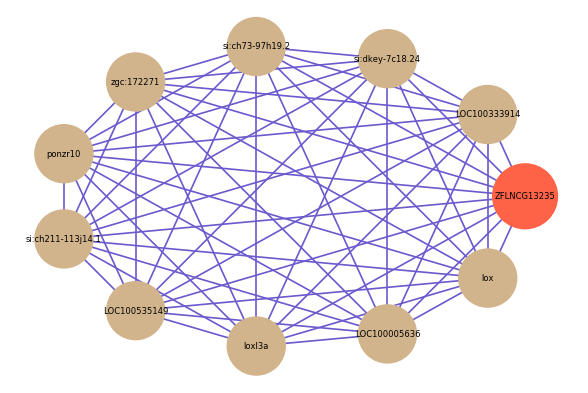
| gene | correlation coefficent |
|---|---|
| LOC100333914 | 0.60 |
| si:dkey-7c18.24 | 0.58 |
| si:ch73-97h19.2 | 0.57 |
| zgc:172271 | 0.55 |
| ponzr10 | 0.55 |
| si:ch211-113j14.1 | 0.55 |
| LOC100535149 | 0.55 |
| loxl3a | 0.54 |
| LOC100005636 | 0.54 |
| lox | 0.54 |
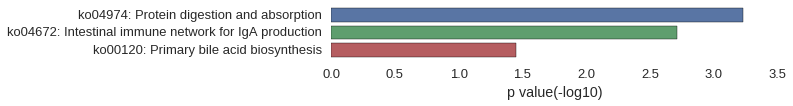
Gene Ontology
Download
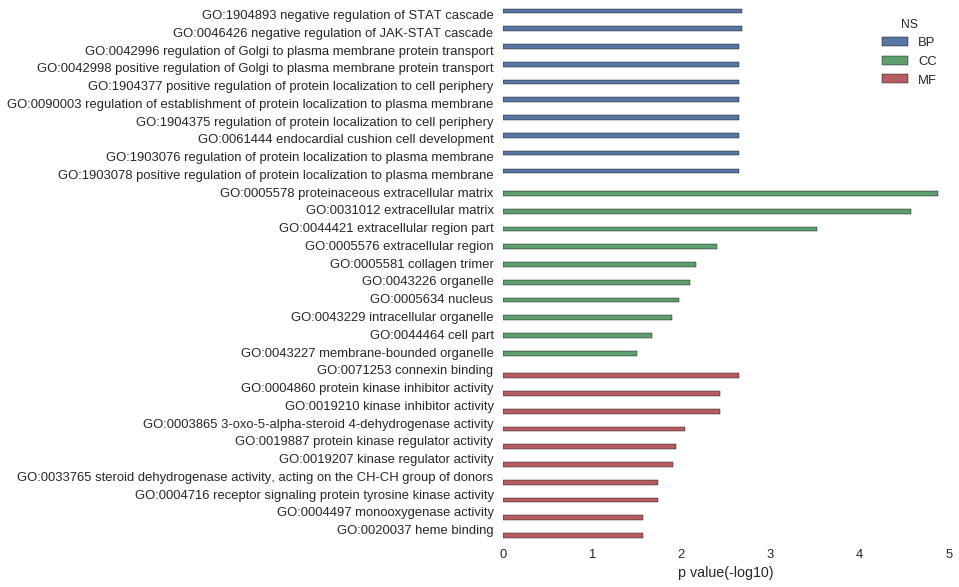
| GO | P value |
|---|---|
| GO:1904893 | 2.09e-03 |
| GO:0046426 | 2.09e-03 |
| GO:0042996 | 2.27e-03 |
| GO:0042998 | 2.27e-03 |
| GO:1904377 | 2.27e-03 |
| GO:0090003 | 2.27e-03 |
| GO:1904375 | 2.27e-03 |
| GO:0061444 | 2.27e-03 |
| GO:1903076 | 2.27e-03 |
| GO:1903078 | 2.27e-03 |
| GO:0005578 | 1.33e-05 |
| GO:0031012 | 2.64e-05 |
| GO:0044421 | 3.00e-04 |
| GO:0005576 | 3.96e-03 |
| GO:0005581 | 6.90e-03 |
| GO:0043226 | 8.04e-03 |
| GO:0005634 | 1.05e-02 |
| GO:0043229 | 1.26e-02 |
| GO:0044464 | 2.13e-02 |
| GO:0043227 | 3.09e-02 |
| GO:0071253 | 2.27e-03 |
| GO:0004860 | 3.70e-03 |
| GO:0019210 | 3.70e-03 |
| GO:0003865 | 9.07e-03 |
| GO:0019887 | 1.13e-02 |
| GO:0019207 | 1.25e-02 |
| GO:0033765 | 1.81e-02 |
| GO:0004716 | 1.81e-02 |
| GO:0004497 | 2.67e-02 |
| GO:0020037 | 2.67e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG13235
TTTAATTCAGCTGTACATGTTTGCCCTATAGGAAGAGCATATGAAAGGCTGAAGAAGCCTGAAAGTGTCC
TCTATTTGAGTGAAGCTCGGGTGTTGATGAAGTTCAGTTGATCAAGGATAATGTTGTGCTGGAACTGCCG
ATATCCGGTGACATTGAAGACGGGTTGTTCTCCAGGAATTTATATCCATCTTTAAACAGAACCTTGAGGA
TGTCCAGAACGACCAA
TTTAATTCAGCTGTACATGTTTGCCCTATAGGAAGAGCATATGAAAGGCTGAAGAAGCCTGAAAGTGTCC
TCTATTTGAGTGAAGCTCGGGTGTTGATGAAGTTCAGTTGATCAAGGATAATGTTGTGCTGGAACTGCCG
ATATCCGGTGACATTGAAGACGGGTTGTTCTCCAGGAATTTATATCCATCTTTAAACAGAACCTTGAGGA
TGTCCAGAACGACCAA