ZFLNCG13259
Basic Information
Chromesome: chr25
Start: 332617
End: 333057
Transcript: ZFLNCT20588 ZFLNCT20589 ZFLNCT20590
Known as:
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 10.17 |
| SRR800045 | muscle | normal | 6.03 |
| SRR519752 | 7 dpf | VD3 treatment | 4.69 |
| SRR519737 | 2 dpf | VD3 treatment | 3.96 |
| SRR519747 | 6 dpf | VD3 treatment | 3.79 |
| SRR516123 | skin | male and 3.5 year | 3.25 |
| SRR519732 | 7 dpf | FETOH treatment | 3.19 |
| SRR658535 | bud | Gata5 morphant | 3.16 |
| SRR658537 | bud | Gata6 morphant | 3.08 |
| SRR800043 | sphere stage | normal | 2.98 |
Express in tissues
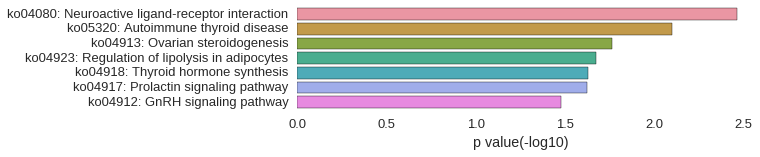
Correlated coding gene
Download
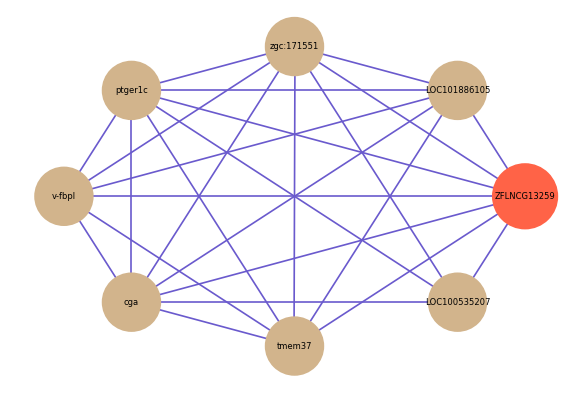
| gene | correlation coefficent |
|---|---|
| LOC101886105 | 0.57 |
| zgc:171551 | 0.54 |
| ptger1c | 0.52 |
| v-fbpl | 0.51 |
| cga | 0.51 |
| tmem37 | 0.51 |
| LOC100535207 | 0.50 |
Gene Ontology
Download
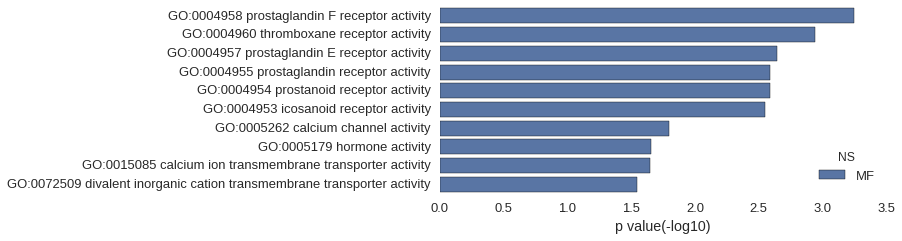
| GO | P value |
|---|---|
| GO:0004958 | 5.68e-04 |
| GO:0004960 | 1.14e-03 |
| GO:0004957 | 2.27e-03 |
| GO:0004955 | 2.56e-03 |
| GO:0004954 | 2.56e-03 |
| GO:0004953 | 2.84e-03 |
| GO:0005262 | 1.61e-02 |
| GO:0005179 | 2.20e-02 |
| GO:0015085 | 2.23e-02 |
| GO:0072509 | 2.84e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG13259
CCAGATCATAGGAACAGGTGTAGATCCAGATCATCAGAACACAAACCGATCCAGATCATCAGAACAGGTG
CAGATCCAGATCATCAGAAAAGGTGTAGATCTAGATCATCAGAACAGGTGCAGTTCCAGATCATCAGAAC
AGGTGCAGATTCAGATCATCAGAACATAAACCGATCCAGATCATCATAACAGGTGCAGATCCAGATCATC
AGAACAGGTGTGGATCCAGATCCATCAGAACACAAACCAATCCAGATCATCAAAATCAATCAGAAAGGTG
TTGATCTGAATAATTAGAACAGGTGTTGATCTGAATAATTAGAACAGGTGTGGATCTAGATCATTAGAAC
AGGTTTAGATTAGAGGTTTAGAATCGTCAGAACAGGTGTGTATATATTTCATCCAAACAGGTGCAGATCC
GAACCGTCAGAAAAGGTTTG
CCAGATCATAGGAACAGGTGTAGATCCAGATCATCAGAACACAAACCGATCCAGATCATCAGAACAGGTG
CAGATCCAGATCATCAGAAAAGGTGTAGATCTAGATCATCAGAACAGGTGCAGTTCCAGATCATCAGAAC
AGGTGCAGATTCAGATCATCAGAACATAAACCGATCCAGATCATCATAACAGGTGCAGATCCAGATCATC
AGAACAGGTGTGGATCCAGATCCATCAGAACACAAACCAATCCAGATCATCAAAATCAATCAGAAAGGTG
TTGATCTGAATAATTAGAACAGGTGTTGATCTGAATAATTAGAACAGGTGTGGATCTAGATCATTAGAAC
AGGTTTAGATTAGAGGTTTAGAATCGTCAGAACAGGTGTGTATATATTTCATCCAAACAGGTGCAGATCC
GAACCGTCAGAAAAGGTTTG