ZFLNCG13573
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR527832 | 16-36 hpf | normal | 18.80 |
| SRR726542 | 5 dpf | infection with control | 9.03 |
| SRR749515 | 24 hpf | eif3ha morphant | 7.24 |
| SRR038627 | embryo | control morpholino | 6.92 |
| SRR633516 | larvae | normal | 6.43 |
| SRR038626 | embryo | control morpholino and bacterial infection | 6.32 |
| SRR633544 | larvae | exposed to cold stress | 5.27 |
| SRR527833 | 5 dpf | normal | 4.83 |
| SRR726540 | 5 dpf | normal | 4.65 |
| SRR594771 | endothelium | normal | 4.49 |
Express in tissues
Correlated coding gene
Download
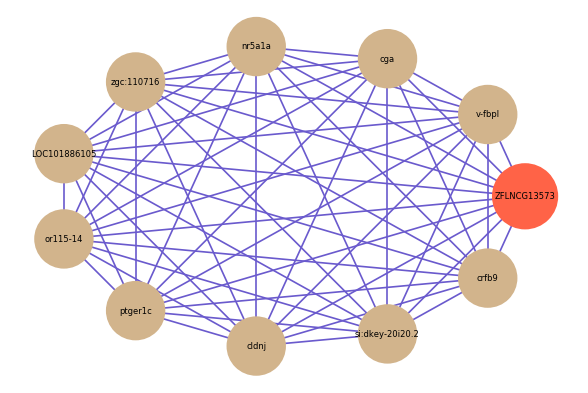
| gene | correlation coefficent |
|---|---|
| v-fbpl | 0.62 |
| cga | 0.61 |
| nr5a1a | 0.60 |
| zgc:110716 | 0.58 |
| LOC101886105 | 0.58 |
| or115-14 | 0.58 |
| ptger1c | 0.58 |
| cldnj | 0.58 |
| si:dkey-20i20.2 | 0.57 |
| crfb9 | 0.57 |
Gene Ontology
Download
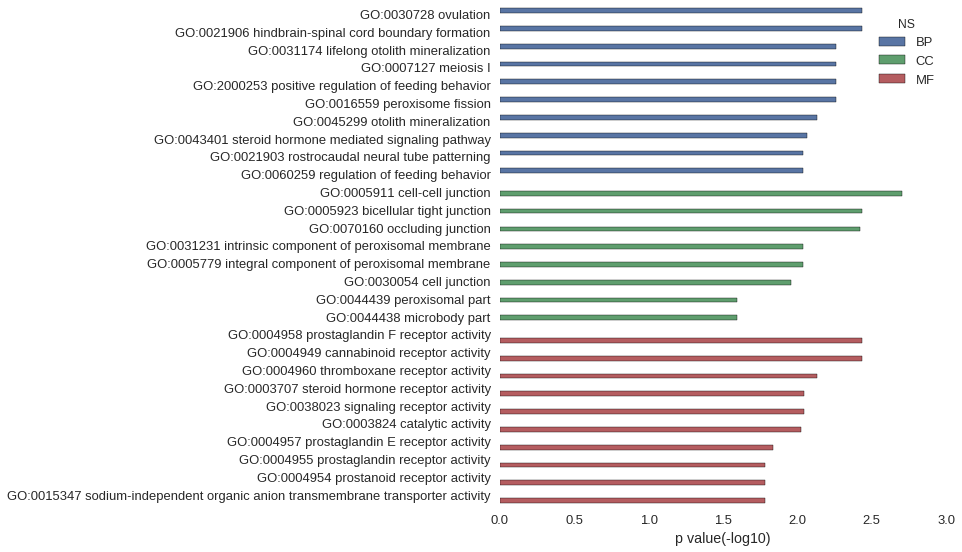
| GO | P value |
|---|---|
| GO:0030728 | 3.69e-03 |
| GO:0021906 | 3.69e-03 |
| GO:0031174 | 5.53e-03 |
| GO:0007127 | 5.53e-03 |
| GO:2000253 | 5.53e-03 |
| GO:0016559 | 5.53e-03 |
| GO:0045299 | 7.37e-03 |
| GO:0043401 | 8.60e-03 |
| GO:0021903 | 9.21e-03 |
| GO:0060259 | 9.21e-03 |
| GO:0005911 | 1.99e-03 |
| GO:0005923 | 3.66e-03 |
| GO:0070160 | 3.81e-03 |
| GO:0031231 | 9.21e-03 |
| GO:0005779 | 9.21e-03 |
| GO:0030054 | 1.10e-02 |
| GO:0044439 | 2.56e-02 |
| GO:0044438 | 2.56e-02 |
| GO:0004958 | 3.69e-03 |
| GO:0004949 | 3.69e-03 |
| GO:0004960 | 7.37e-03 |
| GO:0003707 | 9.04e-03 |
| GO:0038023 | 9.07e-03 |
| GO:0003824 | 9.50e-03 |
| GO:0004957 | 1.47e-02 |
| GO:0004955 | 1.65e-02 |
| GO:0004954 | 1.65e-02 |
| GO:0015347 | 1.65e-02 |
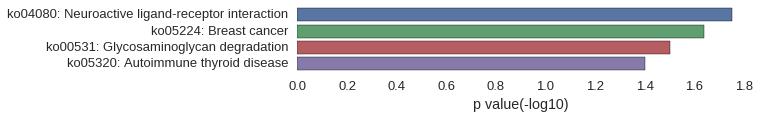
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG13573
ATACTGACTTCAACTGTATATCGCAGAGAAATAAAAGAACGCAATGTCAGATTTTTTCGATATACCCTAC
TTGTAATGCACTTTTCGTACTAAACATTGTCAGATCTGTGTAAGTAGAGCTGCGTGCAGAGCTTCTGATG
ATTTAGTTGTCGTCCACAGCAACGGGAAGAGCTGACGCGGCGGCGGGATAAGATCAGCAGGAAGAGAGAG
CGACTGCTGGCGGACGACCCCGAGGTGGAAAAACACCTTCAGCCAGTGAACGAGGAGCTGGAGTCACTGC
TGGCCAACATCGACTACATCAACGACAGCATCTCCGACTGCCAGGCCAACATCATGCAGATGGAGGAGGC
CAAGGTCACAGCAACACCAGCACCCATTTCACTCGTTTTCCTTCATCTCAGTCCCTTATTAAGCAGAGGT
CACCACAGCAGAATGAACTTGTCACACCTGTTGCAGTGCCCTAGTTTGGACA
ATACTGACTTCAACTGTATATCGCAGAGAAATAAAAGAACGCAATGTCAGATTTTTTCGATATACCCTAC
TTGTAATGCACTTTTCGTACTAAACATTGTCAGATCTGTGTAAGTAGAGCTGCGTGCAGAGCTTCTGATG
ATTTAGTTGTCGTCCACAGCAACGGGAAGAGCTGACGCGGCGGCGGGATAAGATCAGCAGGAAGAGAGAG
CGACTGCTGGCGGACGACCCCGAGGTGGAAAAACACCTTCAGCCAGTGAACGAGGAGCTGGAGTCACTGC
TGGCCAACATCGACTACATCAACGACAGCATCTCCGACTGCCAGGCCAACATCATGCAGATGGAGGAGGC
CAAGGTCACAGCAACACCAGCACCCATTTCACTCGTTTTCCTTCATCTCAGTCCCTTATTAAGCAGAGGT
CACCACAGCAGAATGAACTTGTCACACCTGTTGCAGTGCCCTAGTTTGGACA