ZFLNCG00299
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 7199.15 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 6849.70 |
| SRR1562529 | intestine and pancreas | normal | 3143.69 |
| SRR1004789 | larvae | impdh2 morpholino | 996.71 |
| SRR1004787 | larvae | impdh1a morpholino | 494.26 |
| SRR1035984 | 13 hpf | normal | 419.65 |
| SRR633516 | larvae | normal | 371.08 |
| SRR797908 | embryo | normal | 369.25 |
| SRR797914 | embryo | pbx2/4-MO | 334.85 |
| SRR1004785 | larvae | normal | 329.24 |
Express in tissues
Correlated coding gene
Download
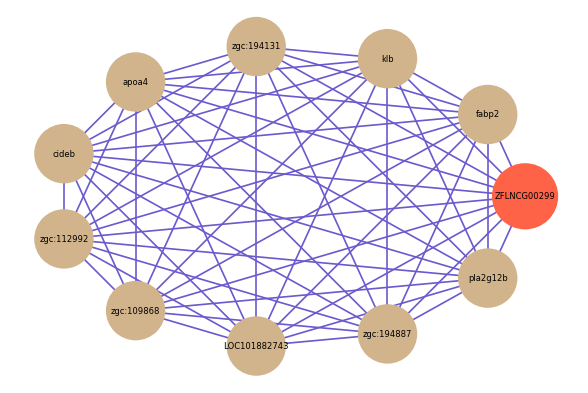
| gene | correlation coefficent |
|---|---|
| fabp2 | 0.90 |
| klb | 0.87 |
| zgc:194131 | 0.86 |
| apoa4 | 0.85 |
| cideb | 0.84 |
| zgc:112992 | 0.84 |
| zgc:109868 | 0.84 |
| LOC101882743 | 0.83 |
| zgc:194887 | 0.83 |
| pla2g12b | 0.83 |
Gene Ontology
Download
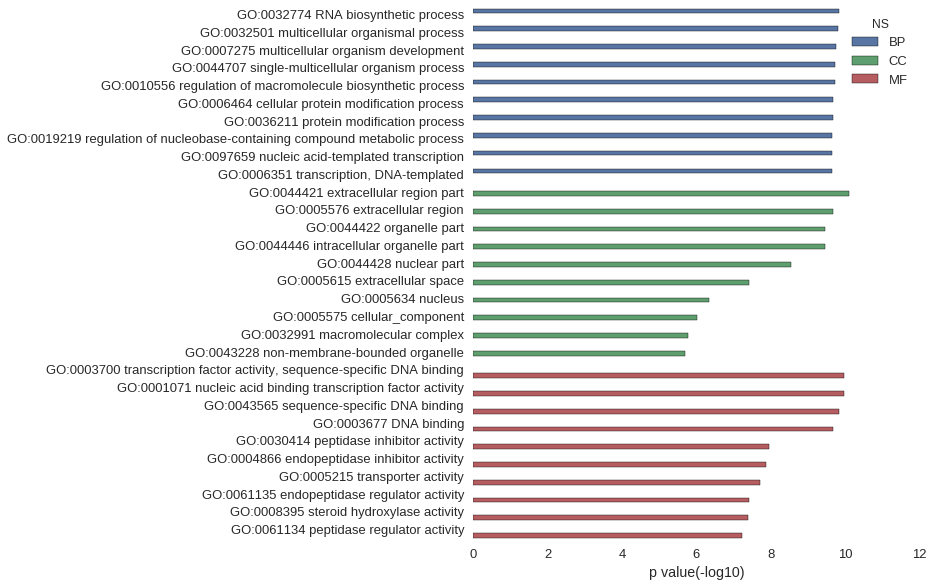
| GO | P value |
|---|---|
| GO:0032774 | 1.46e-10 |
| GO:0032501 | 1.60e-10 |
| GO:0007275 | 1.78e-10 |
| GO:0044707 | 1.84e-10 |
| GO:0010556 | 1.93e-10 |
| GO:0006464 | 2.11e-10 |
| GO:0036211 | 2.11e-10 |
| GO:0019219 | 2.22e-10 |
| GO:0097659 | 2.24e-10 |
| GO:0006351 | 2.24e-10 |
| GO:0044421 | 7.97e-11 |
| GO:0005576 | 2.10e-10 |
| GO:0044422 | 3.46e-10 |
| GO:0044446 | 3.48e-10 |
| GO:0044428 | 2.87e-09 |
| GO:0005615 | 3.80e-08 |
| GO:0005634 | 4.49e-07 |
| GO:0005575 | 9.61e-07 |
| GO:0032991 | 1.73e-06 |
| GO:0043228 | 2.02e-06 |
| GO:0003700 | 1.07e-10 |
| GO:0001071 | 1.07e-10 |
| GO:0043565 | 1.48e-10 |
| GO:0003677 | 2.14e-10 |
| GO:0030414 | 1.15e-08 |
| GO:0004866 | 1.33e-08 |
| GO:0005215 | 1.90e-08 |
| GO:0061135 | 3.93e-08 |
| GO:0008395 | 4.18e-08 |
| GO:0061134 | 5.84e-08 |
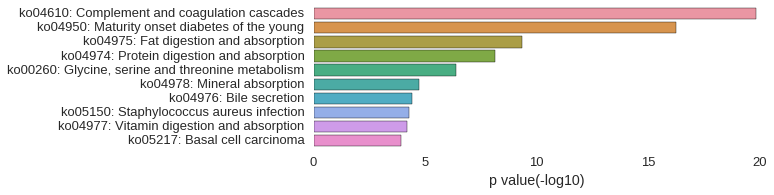
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00299
CTGTCAGGATCACACAACAGTCTGTCATCATCATGACCTTCAACGGGACCTGGAAAGTCGACCGCAATGA
GAACTACGAGAAGTTCATGGAACAAATGGGTAAGTGATGAATAATAATCGATGCTTGCTTGAGTGAGTTG
ATTGATTAAGTGAGATTTACTGCATTCCTTTAACAGGCGTCAACATGGTGAAAAGGAAACTGGCTGCCCA
TGACAACCTGAAGATCACCCTGGAGCAGACCGGAGACAAGTTCAACGTGAAGGAAGTCAGCACTTTCCGC
ACACTGGAAATTAACTTTACTCTGGGCGTCACCTTTGACTATTCTCTGGCAGACGGCACTGAGCTCACAG
TAAGCACAGTCATTGCTGATTTCAGATGCATAAGCGTGGTTCCATAGTAAGCCCATAACATTCAATCTGC
ACAAAGGGTTCTTTATAGTGTGCTGACTGCACTTTTTTTTAAGAATTTTTATTTTAAACATTGTTATACA
AATTGACAAAGGTTTAATTTTTCAGTTTAGCAAAAAATAAAAATAGTAAATTAAATAAATTAAAAGAAGT
TAATACAAAAAACATCAAGAATAATATAGATTCAAAATAACTTAACACACTTTGAGAGAA
CTGTCAGGATCACACAACAGTCTGTCATCATCATGACCTTCAACGGGACCTGGAAAGTCGACCGCAATGA
GAACTACGAGAAGTTCATGGAACAAATGGGTAAGTGATGAATAATAATCGATGCTTGCTTGAGTGAGTTG
ATTGATTAAGTGAGATTTACTGCATTCCTTTAACAGGCGTCAACATGGTGAAAAGGAAACTGGCTGCCCA
TGACAACCTGAAGATCACCCTGGAGCAGACCGGAGACAAGTTCAACGTGAAGGAAGTCAGCACTTTCCGC
ACACTGGAAATTAACTTTACTCTGGGCGTCACCTTTGACTATTCTCTGGCAGACGGCACTGAGCTCACAG
TAAGCACAGTCATTGCTGATTTCAGATGCATAAGCGTGGTTCCATAGTAAGCCCATAACATTCAATCTGC
ACAAAGGGTTCTTTATAGTGTGCTGACTGCACTTTTTTTTAAGAATTTTTATTTTAAACATTGTTATACA
AATTGACAAAGGTTTAATTTTTCAGTTTAGCAAAAAATAAAAATAGTAAATTAAATAAATTAAAAGAAGT
TAATACAAAAAACATCAAGAATAATATAGATTCAAAATAACTTAACACACTTTGAGAGAA