ZFLNCG00614
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 939.99 |
| SRR592701 | pineal gland | normal | 766.64 |
| SRR592702 | pineal gland | normal | 588.37 |
| ERR023144 | brain | normal | 420.40 |
| SRR1028004 | head | normal | 358.65 |
| SRR592699 | pineal gland | normal | 318.61 |
| SRR1028002 | head | normal | 306.70 |
| SRR1648854 | brain | normal | 292.21 |
| SRR592698 | pineal gland | normal | 251.63 |
| SRR1648855 | brain | normal | 238.77 |
Express in tissues
Correlated coding gene
Download
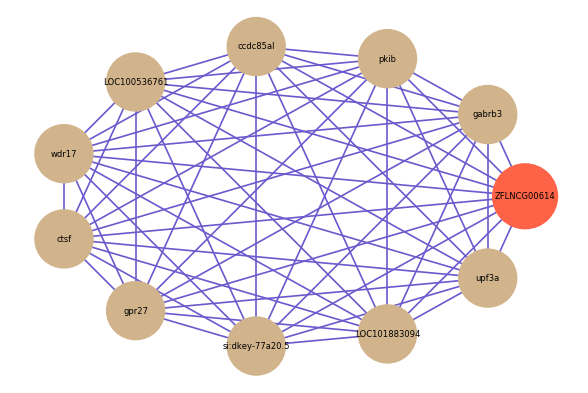
| gene | correlation coefficent |
|---|---|
| gabrb3 | 0.86 |
| pkib | 0.85 |
| ccdc85al | 0.85 |
| LOC100536761 | 0.85 |
| wdr17 | 0.84 |
| ctsf | 0.84 |
| gpr27 | 0.84 |
| si:dkey-77a20.5 | 0.84 |
| LOC101883094 | 0.84 |
| upf3a | 0.84 |
Gene Ontology
Download
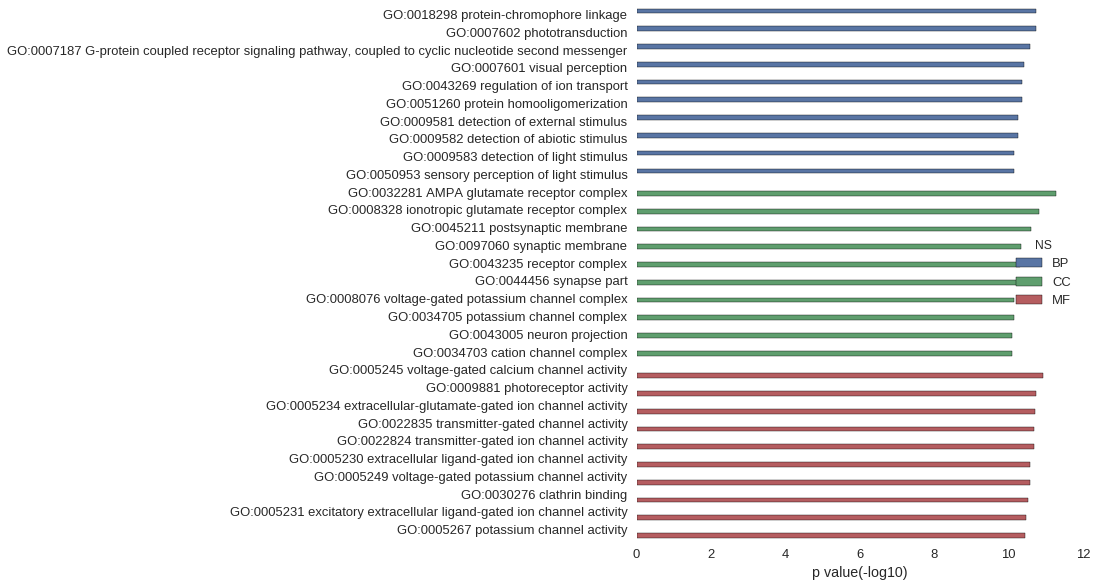
| GO | P value |
|---|---|
| GO:0018298 | 1.86e-11 |
| GO:0007602 | 1.86e-11 |
| GO:0007187 | 2.65e-11 |
| GO:0007601 | 3.82e-11 |
| GO:0043269 | 4.26e-11 |
| GO:0051260 | 4.45e-11 |
| GO:0009581 | 5.73e-11 |
| GO:0009582 | 5.73e-11 |
| GO:0009583 | 7.01e-11 |
| GO:0050953 | 7.23e-11 |
| GO:0032281 | 5.16e-12 |
| GO:0008328 | 1.52e-11 |
| GO:0045211 | 2.42e-11 |
| GO:0097060 | 4.52e-11 |
| GO:0043235 | 5.07e-11 |
| GO:0044456 | 5.48e-11 |
| GO:0008076 | 7.01e-11 |
| GO:0034705 | 7.01e-11 |
| GO:0043005 | 7.91e-11 |
| GO:0034703 | 8.10e-11 |
| GO:0005245 | 1.17e-11 |
| GO:0009881 | 1.86e-11 |
| GO:0005234 | 1.91e-11 |
| GO:0022835 | 2.12e-11 |
| GO:0022824 | 2.12e-11 |
| GO:0005230 | 2.64e-11 |
| GO:0005249 | 2.67e-11 |
| GO:0030276 | 2.98e-11 |
| GO:0005231 | 3.36e-11 |
| GO:0005267 | 3.59e-11 |
KEGG Pathway
Download
Conservation
OMIM
| OMIM | Phenotype |
|---|---|
| 108600 | Spastic ataxia 1, autosomal dominant |
Sequence Download
>ZFLNCG00614
ATTACATTTGACTAGATATTTTGCATGATATTAGTATTCAGCTTAAAGTCCAATGTAAAGGCTTAACTAG
ATTAATTAGGCAAGTCATTGGACAACAGTAGCCTCGGACAGTGCTTAATTTGTAAACTGCGAGGTCCTGG
AACAGATCAGGGTAACCGATCCGGCATGTCACCAGAGGAGGGAGAGGTGTCCCGGATGAAAGAGAAGAGC
G
ATTACATTTGACTAGATATTTTGCATGATATTAGTATTCAGCTTAAAGTCCAATGTAAAGGCTTAACTAG
ATTAATTAGGCAAGTCATTGGACAACAGTAGCCTCGGACAGTGCTTAATTTGTAAACTGCGAGGTCCTGG
AACAGATCAGGGTAACCGATCCGGCATGTCACCAGAGGAGGGAGAGGTGTCCCGGATGAAAGAGAAGAGC
G