ZFLNCG00617
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR594417 | retina | normal | 37.16 |
| SRR1562528 | eye | normal | 31.37 |
| SRR957181 | heart | normal | 26.01 |
| SRR1205166 | 5dpf | transgenic sqET20 and neomycin treated 3h | 17.23 |
| SRR527834 | head | normal | 16.93 |
| SRR1205172 | 5dpf | transgenic sqET20 and neomycin treated 5h | 15.92 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 15.47 |
| SRR1565811 | brain | normal | 15.28 |
| ERR023144 | brain | normal | 13.03 |
| SRR1565809 | brain | normal | 12.98 |
Express in tissues
Correlated coding gene
Download
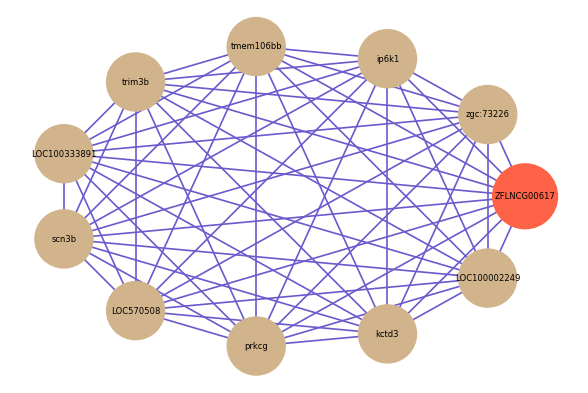
| gene | correlation coefficent |
|---|---|
| zgc:73226 | 0.75 |
| ip6k1 | 0.74 |
| tmem106bb | 0.73 |
| trim3b | 0.71 |
| LOC100333891 | 0.71 |
| scn3b | 0.70 |
| LOC570508 | 0.70 |
| prkcg | 0.70 |
| kctd3 | 0.70 |
| LOC100002249 | 0.70 |
Gene Ontology
Download
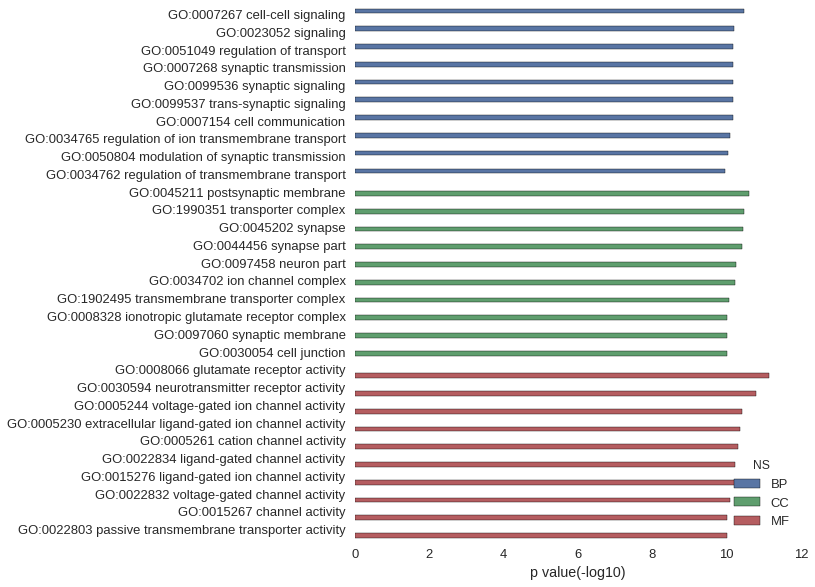
| GO | P value |
|---|---|
| GO:0007267 | 3.49e-11 |
| GO:0023052 | 6.55e-11 |
| GO:0051049 | 6.61e-11 |
| GO:0007268 | 6.71e-11 |
| GO:0099536 | 6.71e-11 |
| GO:0099537 | 6.71e-11 |
| GO:0007154 | 6.76e-11 |
| GO:0034765 | 8.39e-11 |
| GO:0050804 | 9.28e-11 |
| GO:0034762 | 1.14e-10 |
| GO:0045211 | 2.54e-11 |
| GO:1990351 | 3.36e-11 |
| GO:0045202 | 3.62e-11 |
| GO:0044456 | 3.91e-11 |
| GO:0097458 | 5.50e-11 |
| GO:0034702 | 6.09e-11 |
| GO:1902495 | 8.54e-11 |
| GO:0008328 | 9.72e-11 |
| GO:0097060 | 1.00e-10 |
| GO:0030054 | 1.01e-10 |
| GO:0008066 | 7.14e-12 |
| GO:0030594 | 1.64e-11 |
| GO:0005244 | 3.91e-11 |
| GO:0005230 | 4.44e-11 |
| GO:0005261 | 4.93e-11 |
| GO:0022834 | 5.86e-11 |
| GO:0015276 | 5.86e-11 |
| GO:0022832 | 8.27e-11 |
| GO:0015267 | 9.99e-11 |
| GO:0022803 | 9.99e-11 |
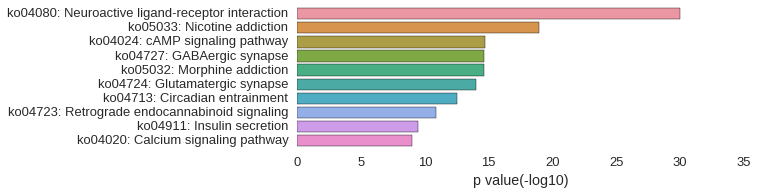
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00617
GAAGAGGAAGAGTTCCGCATGGATATCTGTGGCTCTGACTTATCCAACATCTGATCTCTCCAATCCTCTC
TCTCTTTCACACACACACACACACACACTTTTCTGTCGTTCTCACCATCCACCTGTTTCGCACTCAAACT
GATGAGTTCCAGGCTCAGACATTTACTCAGAAACGCATTACTCTGGATCAAGAAAACGCATTATTTTATC
AATGTGAACGCTTGAGGGATGCGTGACTGAAACCCTTCTGGAGGAACGTCGGCTGATAAACTTATTGGAC
TGTTAATGGGATGAGGAGAGTCATTTTCTGGACGACGGACGACACACTCGAGTAAACCATCAGTTCTTT
GAAGAGGAAGAGTTCCGCATGGATATCTGTGGCTCTGACTTATCCAACATCTGATCTCTCCAATCCTCTC
TCTCTTTCACACACACACACACACACACTTTTCTGTCGTTCTCACCATCCACCTGTTTCGCACTCAAACT
GATGAGTTCCAGGCTCAGACATTTACTCAGAAACGCATTACTCTGGATCAAGAAAACGCATTATTTTATC
AATGTGAACGCTTGAGGGATGCGTGACTGAAACCCTTCTGGAGGAACGTCGGCTGATAAACTTATTGGAC
TGTTAATGGGATGAGGAGAGTCATTTTCTGGACGACGGACGACACACTCGAGTAAACCATCAGTTCTTT