ZFLNCG00784
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562530 | liver | normal | 235.60 |
| SRR1035239 | liver | transgenic mCherry control | 95.64 |
| SRR1562529 | intestine and pancreas | normal | 29.11 |
| SRR1562532 | spleen | normal | 21.67 |
| SRR1035237 | liver | transgenic UHRF1 | 12.10 |
| SRR1647682 | spleen | normal | 10.56 |
| SRR1647681 | head kidney | SVCV treatment | 7.85 |
| SRR726540 | 5 dpf | normal | 7.23 |
| SRR1534349 | larvae | DMSO vehicle | 6.54 |
| SRR1523211 | embryo | control morpholino | 5.47 |
Express in tissues
Correlated coding gene
Download
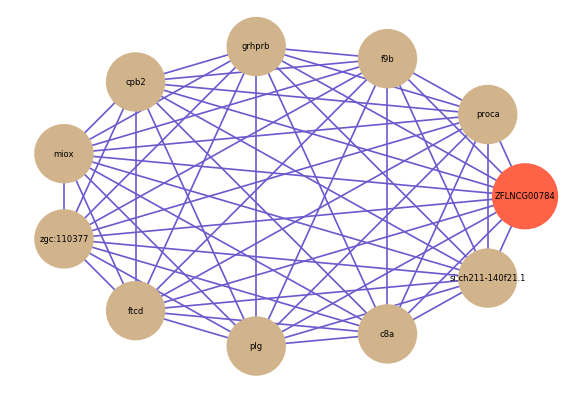
| gene | correlation coefficent |
|---|---|
| proca | 0.80 |
| f9b | 0.76 |
| grhprb | 0.75 |
| cpb2 | 0.74 |
| miox | 0.74 |
| zgc:110377 | 0.73 |
| ftcd | 0.73 |
| plg | 0.73 |
| c8a | 0.73 |
| si:ch211-140f21.1 | 0.73 |
Gene Ontology
Download
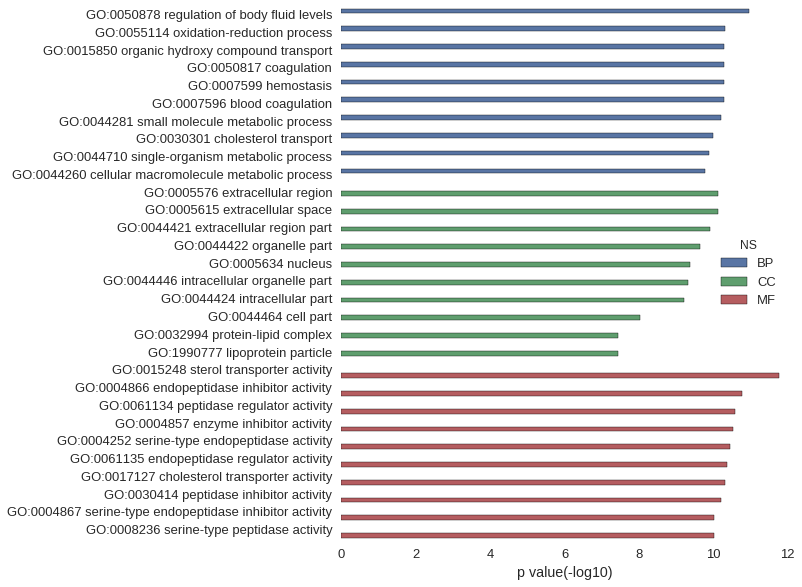
| GO | P value |
|---|---|
| GO:0050878 | 1.07e-11 |
| GO:0055114 | 4.88e-11 |
| GO:0015850 | 5.20e-11 |
| GO:0050817 | 5.22e-11 |
| GO:0007599 | 5.22e-11 |
| GO:0007596 | 5.22e-11 |
| GO:0044281 | 6.15e-11 |
| GO:0030301 | 1.04e-10 |
| GO:0044710 | 1.28e-10 |
| GO:0044260 | 1.65e-10 |
| GO:0005576 | 7.44e-11 |
| GO:0005615 | 7.45e-11 |
| GO:0044421 | 1.25e-10 |
| GO:0044422 | 2.31e-10 |
| GO:0005634 | 4.32e-10 |
| GO:0044446 | 4.66e-10 |
| GO:0044424 | 6.21e-10 |
| GO:0044464 | 9.38e-09 |
| GO:0032994 | 3.75e-08 |
| GO:1990777 | 3.75e-08 |
| GO:0015248 | 1.70e-12 |
| GO:0004866 | 1.65e-11 |
| GO:0061134 | 2.68e-11 |
| GO:0004857 | 2.90e-11 |
| GO:0004252 | 3.58e-11 |
| GO:0061135 | 4.32e-11 |
| GO:0017127 | 4.99e-11 |
| GO:0030414 | 6.24e-11 |
| GO:0004867 | 9.47e-11 |
| GO:0008236 | 9.66e-11 |
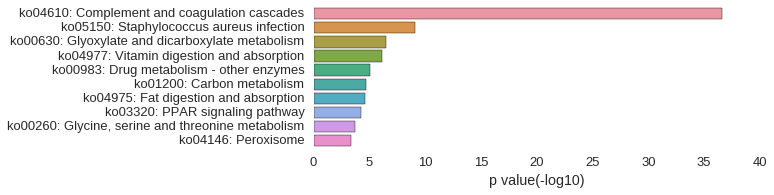
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00784
ATATGTAAAGTGAATAACACATAAAAATGCTCCCGGAGGTCACAATGTTTCCCTTCAAATCCAGAGTCGC
AGGTGCAGGAGAAATCCTGCAACAGGTCAACACACTTGCCATGAACACACGGATTTGTGGTGCACTGGTT
TCCATCTGAAAGTAAAGAAAACCCAATGAGTTATGACGCACTTGCAAAGCTACACCACAGCACAGACTGA
ACTAACCTTTATATGCCGTCCAGAATTCCAGCTGCAGGATGAAGAACTCAGTAAATAGATCATAATTAAG
AGCAAAAGTGCGTAAGAACAGTTAAAAGCTGACCAACCGTGGCCTCCCTTGTTATAAAGATTTCTCTTGC
TTCTTCGAAATCGCACAGTTCCTCTCTGCACTCCCGCTCCAGCGATGCCGGCTTGAGTTCCTCCATGAAA
GTGTTGGCTCTTCTTGAACGGAGCAGCATATGTGCGTTTGGGCTGCTAGAAAATACTGACCAAAACAGAC
ACAGGGTTAATAGGGGAGAGCACTCAATGTGGGTTTCCCAGTATTTATTAATGCTTTAAAAGTTTTCATC
TCCTGTTAATCTTTCATCTCCTGTTAATCTTGCAGTTTATTTGTAAATTTCAGGTAATGGTTCAAATGCG
AGTAATATAGAGCTGTGCCAATAAAATCTGTTTAAAATTGAATACATGTATATAAATAAAATTTATTAAA
GTACATTTAAAATTTCTAAACATAATAGCTCCTCTTATACATGGCTGTTTTTATTGACCTCATACCTGAC
TTGCAAAGAGCCCCATCAGAACAAAAAACAACCAGAGTTAAGGCACAGAAGAAAAGACTCCTCATTATGG
CAACAGACCCTGTAAATACAGCAAAAAATCTATATATTTAAATAATTAAACTTGAAATCAGCATCTAGAT
TTGATGTATGGTAAGTGATGTTTTGGTTTCATATACATAAATGATTATTAACATTAAAGTGAGAATTTTC
ACTGTCTAAAATAATAGCACCTTAAAAAAACTTCTTACCGTGCTCCCCAATATGAACGTGGTACTTTAAA
GGCCAGATTTTTCCTCAAATGAGAACTACATTTGAAGATACATGAAGCAGAAATAAGGATTTTAAATGTC
TCAGCTTAATCCGACAGGACAGCAAAGCACAAACAGCCTTTAGCAGCTAAAGCAAACCGTTCACT
ATATGTAAAGTGAATAACACATAAAAATGCTCCCGGAGGTCACAATGTTTCCCTTCAAATCCAGAGTCGC
AGGTGCAGGAGAAATCCTGCAACAGGTCAACACACTTGCCATGAACACACGGATTTGTGGTGCACTGGTT
TCCATCTGAAAGTAAAGAAAACCCAATGAGTTATGACGCACTTGCAAAGCTACACCACAGCACAGACTGA
ACTAACCTTTATATGCCGTCCAGAATTCCAGCTGCAGGATGAAGAACTCAGTAAATAGATCATAATTAAG
AGCAAAAGTGCGTAAGAACAGTTAAAAGCTGACCAACCGTGGCCTCCCTTGTTATAAAGATTTCTCTTGC
TTCTTCGAAATCGCACAGTTCCTCTCTGCACTCCCGCTCCAGCGATGCCGGCTTGAGTTCCTCCATGAAA
GTGTTGGCTCTTCTTGAACGGAGCAGCATATGTGCGTTTGGGCTGCTAGAAAATACTGACCAAAACAGAC
ACAGGGTTAATAGGGGAGAGCACTCAATGTGGGTTTCCCAGTATTTATTAATGCTTTAAAAGTTTTCATC
TCCTGTTAATCTTTCATCTCCTGTTAATCTTGCAGTTTATTTGTAAATTTCAGGTAATGGTTCAAATGCG
AGTAATATAGAGCTGTGCCAATAAAATCTGTTTAAAATTGAATACATGTATATAAATAAAATTTATTAAA
GTACATTTAAAATTTCTAAACATAATAGCTCCTCTTATACATGGCTGTTTTTATTGACCTCATACCTGAC
TTGCAAAGAGCCCCATCAGAACAAAAAACAACCAGAGTTAAGGCACAGAAGAAAAGACTCCTCATTATGG
CAACAGACCCTGTAAATACAGCAAAAAATCTATATATTTAAATAATTAAACTTGAAATCAGCATCTAGAT
TTGATGTATGGTAAGTGATGTTTTGGTTTCATATACATAAATGATTATTAACATTAAAGTGAGAATTTTC
ACTGTCTAAAATAATAGCACCTTAAAAAAACTTCTTACCGTGCTCCCCAATATGAACGTGGTACTTTAAA
GGCCAGATTTTTCCTCAAATGAGAACTACATTTGAAGATACATGAAGCAGAAATAAGGATTTTAAATGTC
TCAGCTTAATCCGACAGGACAGCAAAGCACAAACAGCCTTTAGCAGCTAAAGCAAACCGTTCACT