ZFLNCG01136
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648856 | brain | normal | 19.47 |
| SRR891511 | brain | normal | 10.71 |
| ERR023144 | brain | normal | 10.01 |
| SRR1648854 | brain | normal | 9.26 |
| SRR1648855 | brain | normal | 5.90 |
| SRR592700 | pineal gland | normal | 4.04 |
| SRR1291414 | 5 dpi | normal | 3.62 |
| ERR023145 | heart | normal | 3.22 |
| SRR372802 | 5 dpf | normal | 3.01 |
| SRR535986 | larvae | nhsl1b mut | 2.89 |
Express in tissues
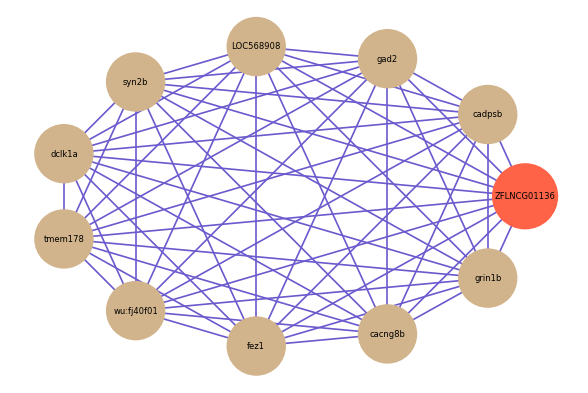
Correlated coding gene
Download
Gene Ontology
Download
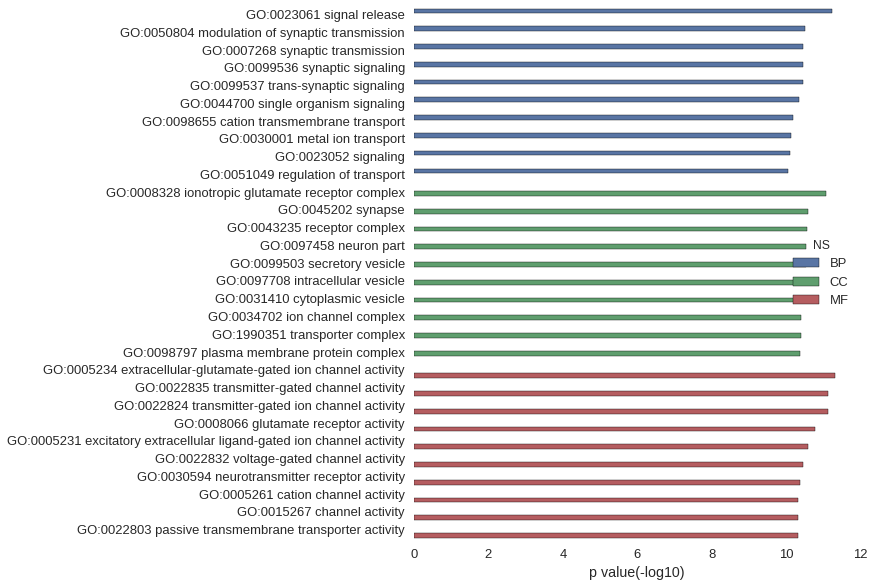
| GO | P value |
|---|---|
| GO:0023061 | 5.78e-12 |
| GO:0050804 | 3.12e-11 |
| GO:0007268 | 3.45e-11 |
| GO:0099536 | 3.45e-11 |
| GO:0099537 | 3.45e-11 |
| GO:0044700 | 4.41e-11 |
| GO:0098655 | 6.43e-11 |
| GO:0030001 | 7.25e-11 |
| GO:0023052 | 7.61e-11 |
| GO:0051049 | 8.68e-11 |
| GO:0008328 | 8.36e-12 |
| GO:0045202 | 2.50e-11 |
| GO:0043235 | 2.68e-11 |
| GO:0097458 | 2.96e-11 |
| GO:0099503 | 2.98e-11 |
| GO:0097708 | 3.19e-11 |
| GO:0031410 | 3.19e-11 |
| GO:0034702 | 3.91e-11 |
| GO:1990351 | 3.92e-11 |
| GO:0098797 | 4.16e-11 |
| GO:0005234 | 4.79e-12 |
| GO:0022835 | 7.54e-12 |
| GO:0022824 | 7.54e-12 |
| GO:0008066 | 1.70e-11 |
| GO:0005231 | 2.54e-11 |
| GO:0022832 | 3.56e-11 |
| GO:0030594 | 4.31e-11 |
| GO:0005261 | 4.65e-11 |
| GO:0015267 | 4.83e-11 |
| GO:0022803 | 4.83e-11 |
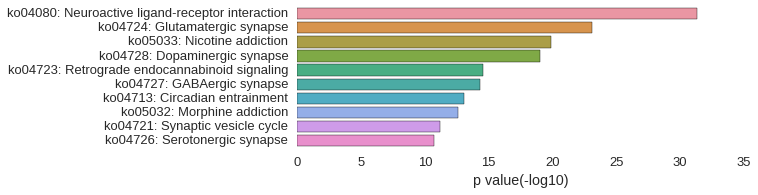
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01136
GAAACTCTACTGAAACTCTCTTTAGTGCATTCGAGTTCACACTTGATGGAGCGACTGCCTCTCAGACACA
CATTTTATCATCTCTGCCCAGTGTGCCATGGGGGCTTTGTGTAGTTCTGACAGACTGTATGCCAAAGTGT
GAGCCAAGCCAAGGCTAGCAGATGCTAACTGGATGAGCTGAACATAACTGAAATCCAGACCCCCTGTATG
AGTGTCTCTCTCCTTTCTCTTGCCCTCTCTGTTTTTCTAATTATCCAAGTTCCTCTTGTTAAAAAAAAAA
AAACTGTGGTCACACCATGACACAGTAATGGTGTTG
GAAACTCTACTGAAACTCTCTTTAGTGCATTCGAGTTCACACTTGATGGAGCGACTGCCTCTCAGACACA
CATTTTATCATCTCTGCCCAGTGTGCCATGGGGGCTTTGTGTAGTTCTGACAGACTGTATGCCAAAGTGT
GAGCCAAGCCAAGGCTAGCAGATGCTAACTGGATGAGCTGAACATAACTGAAATCCAGACCCCCTGTATG
AGTGTCTCTCTCCTTTCTCTTGCCCTCTCTGTTTTTCTAATTATCCAAGTTCCTCTTGTTAAAAAAAAAA
AAACTGTGGTCACACCATGACACAGTAATGGTGTTG