ZFLNCG01253
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800045 | muscle | normal | 479.63 |
| SRR800043 | sphere stage | normal | 395.45 |
| SRR800046 | sphere stage | 5azaCyD treatment | 326.48 |
| SRR800049 | sphere stage | control treatment | 252.28 |
| SRR800037 | egg | normal | 143.37 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 68.75 |
| SRR1021215 | sphere stage | Mzeomesa | 48.09 |
| SRR1021213 | sphere stage | normal | 33.81 |
| SRR658537 | bud | Gata6 morphant | 30.94 |
| SRR748491 | germ ring | normal | 21.61 |
Express in tissues
Correlated coding gene
Download
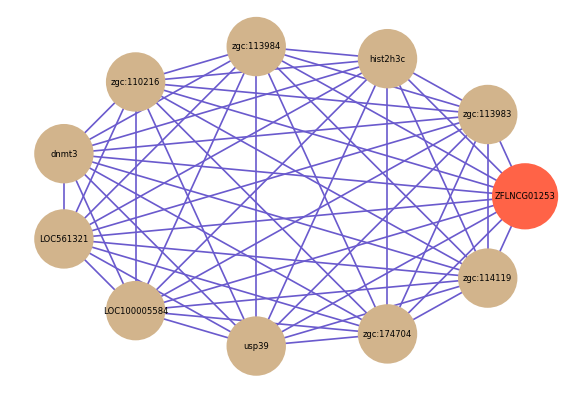
| gene | correlation coefficent |
|---|---|
| zgc:113983 | 0.72 |
| hist2h3c | 0.70 |
| zgc:113984 | 0.70 |
| zgc:110216 | 0.69 |
| dnmt3 | 0.69 |
| LOC561321 | 0.68 |
| LOC100005584 | 0.68 |
| usp39 | 0.68 |
| zgc:174704 | 0.67 |
| zgc:114119 | 0.67 |
Gene Ontology
Download
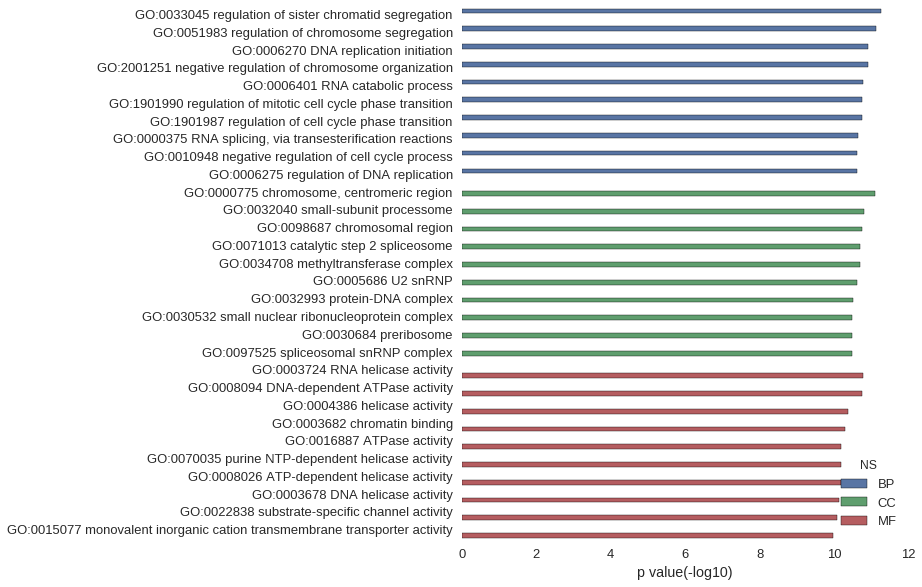
| GO | P value |
|---|---|
| GO:0033045 | 5.59e-12 |
| GO:0051983 | 7.50e-12 |
| GO:0006270 | 1.24e-11 |
| GO:2001251 | 1.24e-11 |
| GO:0006401 | 1.62e-11 |
| GO:1901990 | 1.76e-11 |
| GO:1901987 | 1.76e-11 |
| GO:0000375 | 2.19e-11 |
| GO:0010948 | 2.34e-11 |
| GO:0006275 | 2.43e-11 |
| GO:0000775 | 7.67e-12 |
| GO:0032040 | 1.54e-11 |
| GO:0098687 | 1.76e-11 |
| GO:0071013 | 1.95e-11 |
| GO:0034708 | 1.95e-11 |
| GO:0005686 | 2.43e-11 |
| GO:0032993 | 3.01e-11 |
| GO:0030532 | 3.22e-11 |
| GO:0030684 | 3.22e-11 |
| GO:0097525 | 3.22e-11 |
| GO:0003724 | 1.62e-11 |
| GO:0008094 | 1.76e-11 |
| GO:0004386 | 4.22e-11 |
| GO:0003682 | 5.01e-11 |
| GO:0016887 | 6.45e-11 |
| GO:0070035 | 6.54e-11 |
| GO:0008026 | 6.54e-11 |
| GO:0003678 | 7.21e-11 |
| GO:0022838 | 8.02e-11 |
| GO:0015077 | 1.04e-10 |
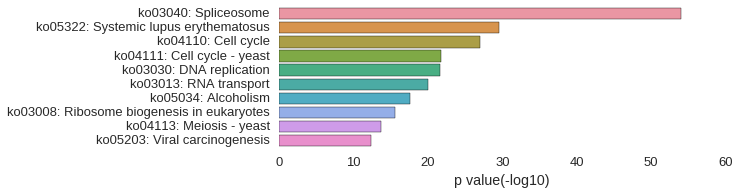
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01253
GCCCTCAGATTACCAAAGAAGCAGCTCATAGCTCATTGGACATTGCAGAGAACCAAGTTTCAACCTGTGT
CCACACACACTCAAACATACACACAAATGACCGTTCTTCCCATCTCTCTCGAACATTCCTCTGACTAAGA
GCTTGATGAGAAAAACTTGTTTAATGTAGGCAGGCCCTTACAGCAAAGCAGCAAATCTCCGCTGCCCAGC
CAAGATTACCAGCCAATAGGAGCACAGGAGAGCTCGGACTCTTATCCACTGAGCTCGCATTCGCCTTGGT
CTCATCATTGGGC
GCCCTCAGATTACCAAAGAAGCAGCTCATAGCTCATTGGACATTGCAGAGAACCAAGTTTCAACCTGTGT
CCACACACACTCAAACATACACACAAATGACCGTTCTTCCCATCTCTCTCGAACATTCCTCTGACTAAGA
GCTTGATGAGAAAAACTTGTTTAATGTAGGCAGGCCCTTACAGCAAAGCAGCAAATCTCCGCTGCCCAGC
CAAGATTACCAGCCAATAGGAGCACAGGAGAGCTCGGACTCTTATCCACTGAGCTCGCATTCGCCTTGGT
CTCATCATTGGGC