ZFLNCG01450
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891511 | brain | normal | 31.11 |
| ERR023144 | brain | normal | 22.86 |
| SRR1565809 | brain | normal | 13.16 |
| SRR1565805 | brain | normal | 12.70 |
| SRR1565819 | brain | normal | 12.65 |
| SRR630463 | 5 dpf | normal | 11.96 |
| SRR1565815 | brain | normal | 11.85 |
| SRR1565811 | brain | normal | 11.75 |
| SRR1565817 | brain | normal | 11.52 |
| SRR1565807 | brain | normal | 11.41 |
Express in tissues
Correlated coding gene
Download
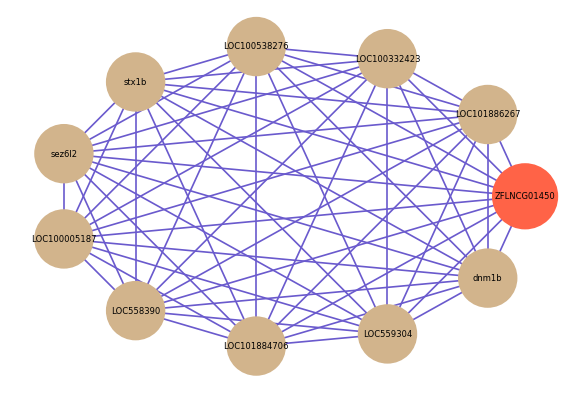
| gene | correlation coefficent |
|---|---|
| LOC101886267 | 0.87 |
| LOC100332423 | 0.86 |
| LOC100538276 | 0.85 |
| stx1b | 0.85 |
| sez6l2 | 0.85 |
| LOC100005187 | 0.85 |
| LOC558390 | 0.85 |
| LOC101884706 | 0.85 |
| LOC559304 | 0.85 |
| dnm1b | 0.85 |
Gene Ontology
Download
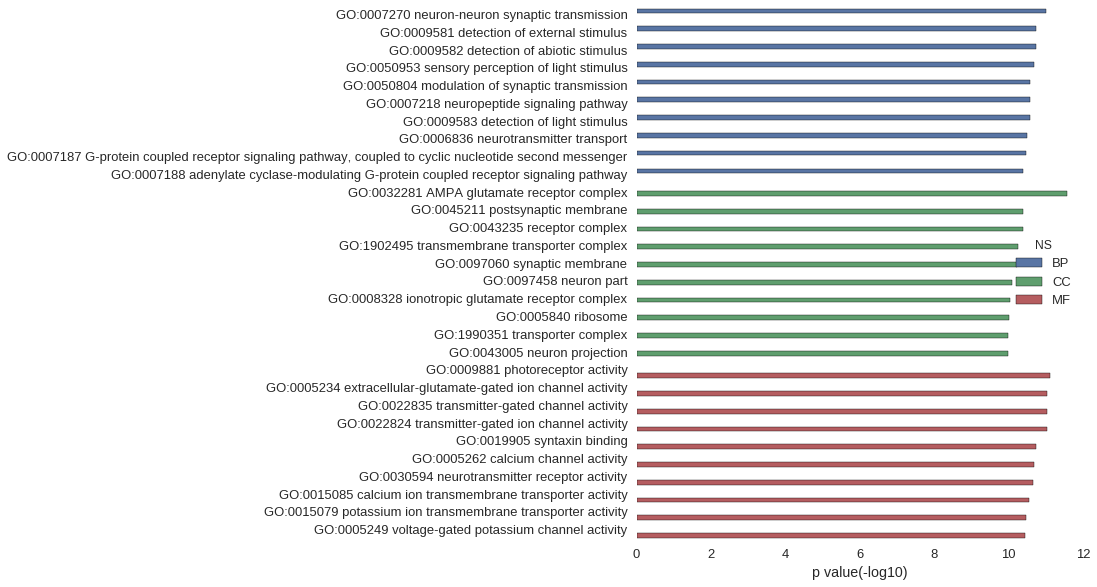
| GO | P value |
|---|---|
| GO:0007270 | 9.90e-12 |
| GO:0009581 | 1.78e-11 |
| GO:0009582 | 1.78e-11 |
| GO:0050953 | 2.12e-11 |
| GO:0050804 | 2.62e-11 |
| GO:0007218 | 2.66e-11 |
| GO:0009583 | 2.71e-11 |
| GO:0006836 | 3.13e-11 |
| GO:0007187 | 3.44e-11 |
| GO:0007188 | 3.98e-11 |
| GO:0032281 | 2.62e-12 |
| GO:0045211 | 4.02e-11 |
| GO:0043235 | 4.14e-11 |
| GO:1902495 | 5.56e-11 |
| GO:0097060 | 5.76e-11 |
| GO:0097458 | 8.18e-11 |
| GO:0008328 | 9.30e-11 |
| GO:0005840 | 9.95e-11 |
| GO:1990351 | 1.01e-10 |
| GO:0043005 | 1.05e-10 |
| GO:0009881 | 7.88e-12 |
| GO:0005234 | 9.00e-12 |
| GO:0022835 | 9.31e-12 |
| GO:0022824 | 9.31e-12 |
| GO:0019905 | 1.78e-11 |
| GO:0005262 | 2.12e-11 |
| GO:0030594 | 2.20e-11 |
| GO:0015085 | 2.90e-11 |
| GO:0015079 | 3.47e-11 |
| GO:0005249 | 3.67e-11 |
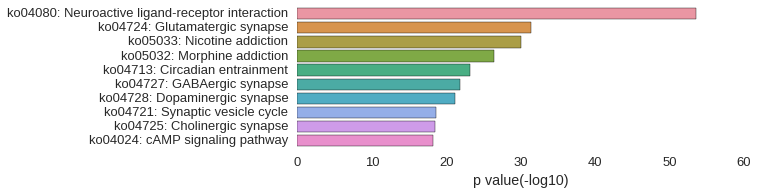
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01450
AAGAACACACACCCTAAACACTGTGATGATAATAATAATCGTAGTAGTAGTAATAATAGTAGGAGTAATA
ATAATAATAATAATGGTGTGAACAGAAAAGGAGACGAGAGGAGATGAGCGGTGGACGCACCTCGCGATAG
CTTCACCAGTAAAAACAGCACACGATATCAGCTCACTAATTAGCTTTACAAAGTCAGTCAGTCCTTAGAA
AGGGTGAAAAAGCCCTGATCGAGACACACACTCACTGCGTGTGTGTGTGTGTGTTTACTTAGCTACACCT
TTGAGGACTCGTTTATTAGTTGAAACCTTCACTTGTAAACATTCCCACAGACGCACTTATCGTGAATGCG
TCTCGGTTTGAGGCATAAGGGAGATGACCGTTTTCAAGCTTTACCAAGGTTTAGAAAAGTCAAGATTTTA
AACTGTCAATATTCCGTCAATATTAAGGTTCCCGTCAATATTTCGTCAATATTAAATATTCTGTCAATAT
TAAGGTTTCCGTCAATATTCTGTCAATATTAAATGTTCCATCAATATTCCCTCTATATTAAGGTTTCCGT
AAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCCTCTGTATTAAAGTTTCCGTCAATATTCCGTC
AATATTAAATGTTCCGTCAATATTCCGTCTATATTAAGGTTTCCATCAATATTCTGTCAATATTAAATGT
TCCGTCAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCGTCTATAATAAGGTTTCCGTCAATATT
CCGTCAATATTAAATATTCCGTCAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCGTCTATAATA
AGGTTTCCGTCAATATTCCGTCAATATTAAATATTCCGTCAATATTCCGTCAATATTAAATGTTCCGTCA
ATATTCCGTCTATAATAAGGTTTCCGTCAATATTCCGTCAATATTAAATATTCCGTCAATATTCCGTCTA
TATTAAGGTTTCTGTCAATATTCTGTCAATATTAAATGTTCCGTCAATATTCCGTCAATATTAAATGTTC
CGTCAATATTCCGTCTATAATAAGGTTTCCGTCAATATTCCGTCAATATTAAATATTCCGTCAATATTCC
GTCTATATTAAGGTTTCTGTCAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCC
AAGAACACACACCCTAAACACTGTGATGATAATAATAATCGTAGTAGTAGTAATAATAGTAGGAGTAATA
ATAATAATAATAATGGTGTGAACAGAAAAGGAGACGAGAGGAGATGAGCGGTGGACGCACCTCGCGATAG
CTTCACCAGTAAAAACAGCACACGATATCAGCTCACTAATTAGCTTTACAAAGTCAGTCAGTCCTTAGAA
AGGGTGAAAAAGCCCTGATCGAGACACACACTCACTGCGTGTGTGTGTGTGTGTTTACTTAGCTACACCT
TTGAGGACTCGTTTATTAGTTGAAACCTTCACTTGTAAACATTCCCACAGACGCACTTATCGTGAATGCG
TCTCGGTTTGAGGCATAAGGGAGATGACCGTTTTCAAGCTTTACCAAGGTTTAGAAAAGTCAAGATTTTA
AACTGTCAATATTCCGTCAATATTAAGGTTCCCGTCAATATTTCGTCAATATTAAATATTCTGTCAATAT
TAAGGTTTCCGTCAATATTCTGTCAATATTAAATGTTCCATCAATATTCCCTCTATATTAAGGTTTCCGT
AAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCCTCTGTATTAAAGTTTCCGTCAATATTCCGTC
AATATTAAATGTTCCGTCAATATTCCGTCTATATTAAGGTTTCCATCAATATTCTGTCAATATTAAATGT
TCCGTCAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCGTCTATAATAAGGTTTCCGTCAATATT
CCGTCAATATTAAATATTCCGTCAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCGTCTATAATA
AGGTTTCCGTCAATATTCCGTCAATATTAAATATTCCGTCAATATTCCGTCAATATTAAATGTTCCGTCA
ATATTCCGTCTATAATAAGGTTTCCGTCAATATTCCGTCAATATTAAATATTCCGTCAATATTCCGTCTA
TATTAAGGTTTCTGTCAATATTCTGTCAATATTAAATGTTCCGTCAATATTCCGTCAATATTAAATGTTC
CGTCAATATTCCGTCTATAATAAGGTTTCCGTCAATATTCCGTCAATATTAAATATTCCGTCAATATTCC
GTCTATATTAAGGTTTCTGTCAATATTCCGTCAATATTAAATGTTCCGTCAATATTCCC