ZFLNCG01968
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 51.39 |
| ERR023145 | heart | normal | 48.70 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 41.02 |
| ERR023146 | head kidney | normal | 34.18 |
| SRR1648856 | brain | normal | 25.33 |
| ERR023144 | brain | normal | 24.30 |
| ERR023143 | swim bladder | normal | 22.30 |
| ERR594417 | retina | normal | 21.30 |
| SRR1562528 | eye | normal | 20.45 |
| ERR594416 | retina | transgenic flk1 | 18.14 |
Express in tissues
Correlated coding gene
Download
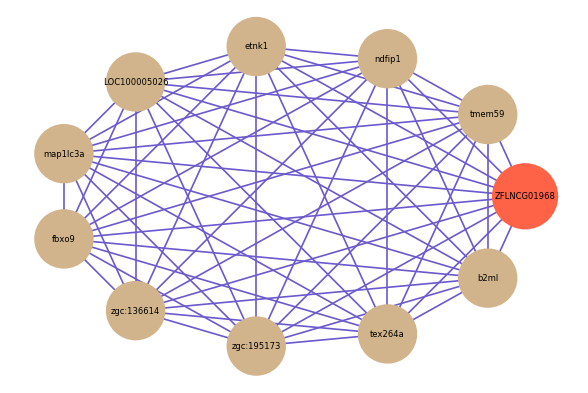
| gene | correlation coefficent |
|---|---|
| tmem59 | 0.85 |
| ndfip1 | 0.84 |
| etnk1 | 0.83 |
| LOC100005026 | 0.83 |
| map1lc3a | 0.83 |
| fbxo9 | 0.82 |
| zgc:136614 | 0.82 |
| zgc:195173 | 0.82 |
| tex264a | 0.82 |
| b2ml | 0.82 |
Gene Ontology
Download
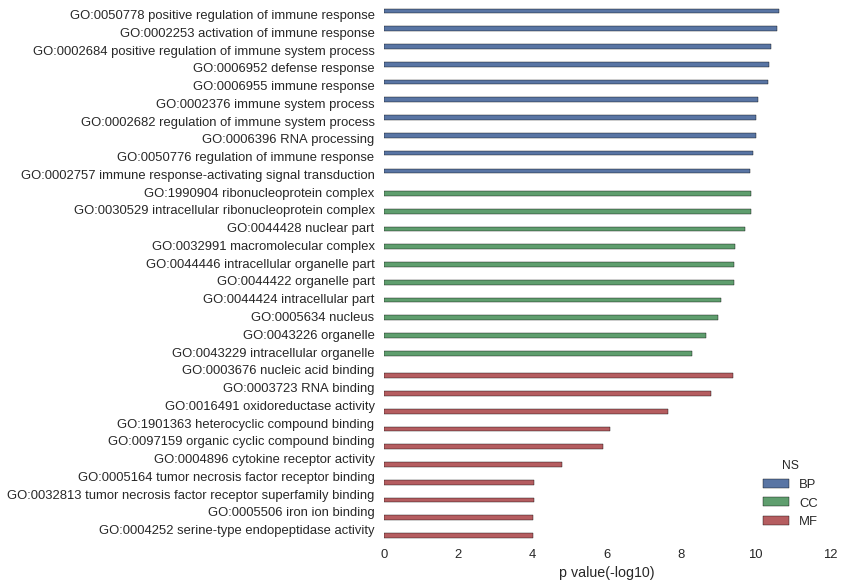
| GO | P value |
|---|---|
| GO:0050778 | 2.42e-11 |
| GO:0002253 | 2.62e-11 |
| GO:0002684 | 3.86e-11 |
| GO:0006952 | 4.50e-11 |
| GO:0006955 | 4.55e-11 |
| GO:0002376 | 8.50e-11 |
| GO:0002682 | 9.52e-11 |
| GO:0006396 | 9.58e-11 |
| GO:0050776 | 1.15e-10 |
| GO:0002757 | 1.44e-10 |
| GO:1990904 | 1.37e-10 |
| GO:0030529 | 1.37e-10 |
| GO:0044428 | 1.92e-10 |
| GO:0032991 | 3.61e-10 |
| GO:0044446 | 3.72e-10 |
| GO:0044422 | 3.90e-10 |
| GO:0044424 | 8.38e-10 |
| GO:0005634 | 1.02e-09 |
| GO:0043226 | 2.21e-09 |
| GO:0043229 | 5.06e-09 |
| GO:0003676 | 4.08e-10 |
| GO:0003723 | 1.62e-09 |
| GO:0016491 | 2.33e-08 |
| GO:1901363 | 8.30e-07 |
| GO:0097159 | 1.24e-06 |
| GO:0004896 | 1.56e-05 |
| GO:0005164 | 9.32e-05 |
| GO:0032813 | 9.32e-05 |
| GO:0005506 | 9.47e-05 |
| GO:0004252 | 9.55e-05 |
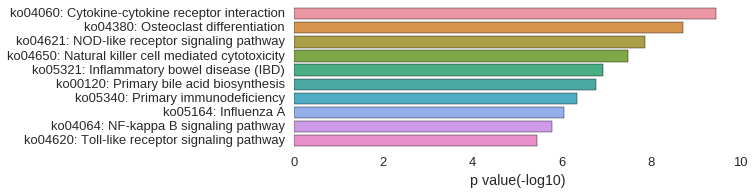
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG01968
TCTTCATATATACGCTTTTATTAGGAGACCCCCAATTAAAGGTGCAAAGGAATAAACTCTGAATCTGATA
TAATTAACGTGGCCTGATTAGGACACTAATATTTCGTTTCAACATTCAAAGGGCTCTTAATATTTACAGC
ACTTGGCCCCTGAATAGGAGTCGATTCAAATCTCTGAATATTAGACTTTAAATAACAGCGAATAATCTTA
TAGAAGAATAAACTTATACAAGGTGCAAATCAAGTAACAGCTTGTTACTTTCTGAATAATACATCATCTG
TACATCACAGTTATGTTCGGTGACAAACAATCGGTCGAGTTATAAATACACAATCGAACAAAATGCATGG
AGAAAACTTAGTTACAGCATTTTCGTTAGCCTGTGCTGGATCGACACCCCTTTAGATGTACGACAACAAC
AAAAAGAAAATAAAAAATGCATCTCCTTATTTACAGTTGCGATTCTCTTCACAGCTCAATTCCCAAAGGT
AAAACACTGCAAGAGAGTGACACGAACAACACAACGTGACCGTCGACAGCTGGTCACTTCTGCTGCCGCT
CGCTCACTCACTCGCAGTGCTTATGTTGTGTAACGTACCACATCAAGTACTTTCTTTCACTCTAAACCAA
TCTTCATATATACGCTTTTATTAGGAGACCCCCAATTAAAGGTGCAAAGGAATAAACTCTGAATCTGATA
TAATTAACGTGGCCTGATTAGGACACTAATATTTCGTTTCAACATTCAAAGGGCTCTTAATATTTACAGC
ACTTGGCCCCTGAATAGGAGTCGATTCAAATCTCTGAATATTAGACTTTAAATAACAGCGAATAATCTTA
TAGAAGAATAAACTTATACAAGGTGCAAATCAAGTAACAGCTTGTTACTTTCTGAATAATACATCATCTG
TACATCACAGTTATGTTCGGTGACAAACAATCGGTCGAGTTATAAATACACAATCGAACAAAATGCATGG
AGAAAACTTAGTTACAGCATTTTCGTTAGCCTGTGCTGGATCGACACCCCTTTAGATGTACGACAACAAC
AAAAAGAAAATAAAAAATGCATCTCCTTATTTACAGTTGCGATTCTCTTCACAGCTCAATTCCCAAAGGT
AAAACACTGCAAGAGAGTGACACGAACAACACAACGTGACCGTCGACAGCTGGTCACTTCTGCTGCCGCT
CGCTCACTCACTCGCAGTGCTTATGTTGTGTAACGTACCACATCAAGTACTTTCTTTCACTCTAAACCAA