ZFLNCG02048
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 28.42 |
| SRR1648854 | brain | normal | 19.39 |
| SRR1648856 | brain | normal | 17.51 |
| SRR1523211 | embryo | control morpholino | 9.73 |
| SRR1291417 | 5 dpi | injected marinum | 8.94 |
| SRR630463 | 5 dpf | normal | 8.69 |
| SRR630464 | 5 dpf | injected epidermidis | 7.70 |
| SRR1291414 | 5 dpi | normal | 7.63 |
| SRR1523214 | embryo | glut12 morpholino | 7.45 |
| SRR1565807 | brain | normal | 7.44 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
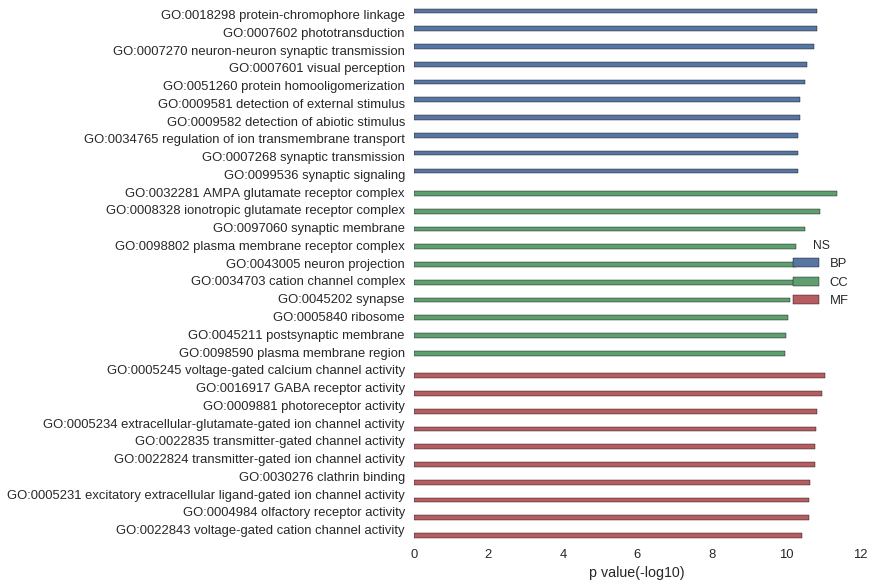
| GO | P value |
|---|---|
| GO:0018298 | 1.47e-11 |
| GO:0007602 | 1.47e-11 |
| GO:0007270 | 1.75e-11 |
| GO:0007601 | 2.71e-11 |
| GO:0051260 | 3.11e-11 |
| GO:0009581 | 4.14e-11 |
| GO:0009582 | 4.14e-11 |
| GO:0034765 | 4.81e-11 |
| GO:0007268 | 4.92e-11 |
| GO:0099536 | 4.92e-11 |
| GO:0032281 | 4.28e-12 |
| GO:0008328 | 1.19e-11 |
| GO:0097060 | 3.07e-11 |
| GO:0098802 | 5.43e-11 |
| GO:0043005 | 5.43e-11 |
| GO:0034703 | 5.60e-11 |
| GO:0045202 | 7.90e-11 |
| GO:0005840 | 8.65e-11 |
| GO:0045211 | 9.90e-11 |
| GO:0098590 | 1.05e-10 |
| GO:0005245 | 9.04e-12 |
| GO:0016917 | 1.06e-11 |
| GO:0009881 | 1.47e-11 |
| GO:0005234 | 1.55e-11 |
| GO:0022835 | 1.69e-11 |
| GO:0022824 | 1.69e-11 |
| GO:0030276 | 2.25e-11 |
| GO:0005231 | 2.41e-11 |
| GO:0004984 | 2.41e-11 |
| GO:0022843 | 3.72e-11 |
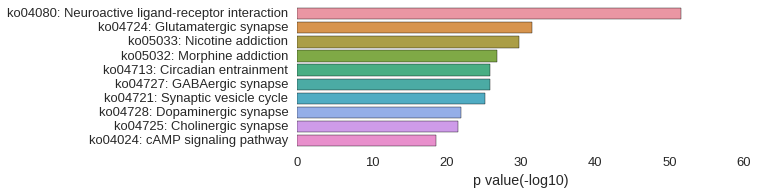
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG02048
CACTGCAGTACATTTCGTTACAACTATTGATAAAAATAAACACAACATATTCTATCAACGTTCACCATTT
ATAAACTCACCTATCATAGCTTTATTCTGTGACGCATAACTATGAGCACCAAATCTAAAAACTTGCACAG
TCACTTGCTCACCAAACGAAAATATCGTAAACTATTAGCCTAAATCTTGATAATGCTACCATTAACCTGC
TTTCTGGAAAACAAGCTTTTGTTCACTAAGGATTTAAAAAAGATAATACGGTAACACTAAACAATACGGT
AAACACTTTACAATAAGGTTCATTAGTTAATGCATTTACTAACATGAACTAATCATGAACAACACATGTA
CAGAATTTATTAATCATAATTGAACATTTACTAGTACATTATTAACATCCAAGTCCATGCTTGTTAAC
CACTGCAGTACATTTCGTTACAACTATTGATAAAAATAAACACAACATATTCTATCAACGTTCACCATTT
ATAAACTCACCTATCATAGCTTTATTCTGTGACGCATAACTATGAGCACCAAATCTAAAAACTTGCACAG
TCACTTGCTCACCAAACGAAAATATCGTAAACTATTAGCCTAAATCTTGATAATGCTACCATTAACCTGC
TTTCTGGAAAACAAGCTTTTGTTCACTAAGGATTTAAAAAAGATAATACGGTAACACTAAACAATACGGT
AAACACTTTACAATAAGGTTCATTAGTTAATGCATTTACTAACATGAACTAATCATGAACAACACATGTA
CAGAATTTATTAATCATAATTGAACATTTACTAGTACATTATTAACATCCAAGTCCATGCTTGTTAAC