ZFLNCG03008
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR519737 | 2 dpf | VD3 treatment | 72.75 |
| SRR519717 | 2 dpf | FETOH treatment | 64.64 |
| SRR519747 | 6 dpf | VD3 treatment | 30.67 |
| SRR519727 | 6 dpf | FETOH treatment | 29.72 |
| SRR519722 | 4 dpf | FETOH treatment | 28.32 |
| SRR519742 | 4 dpf | VD3 treatment | 26.28 |
| SRR519732 | 7 dpf | FETOH treatment | 25.94 |
| SRR519752 | 7 dpf | VD3 treatment | 24.48 |
| SRR038627 | embryo | control morpholino | 18.52 |
| SRR726542 | 5 dpf | infection with control | 18.00 |
Express in tissues
Correlated coding gene
Download
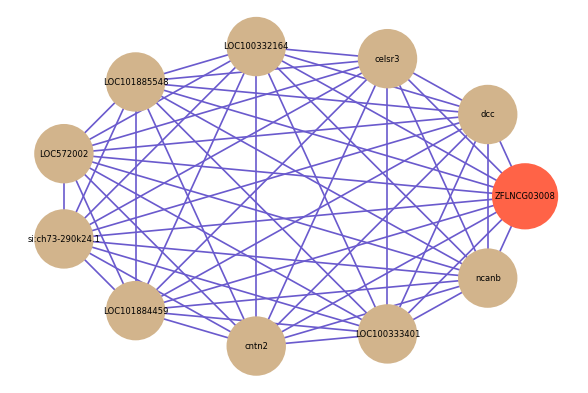
| gene | correlation coefficent |
|---|---|
| dcc | 0.79 |
| celsr3 | 0.79 |
| LOC100332164 | 0.79 |
| LOC101885548 | 0.79 |
| LOC572002 | 0.77 |
| si:ch73-290k24.1 | 0.77 |
| LOC101884459 | 0.76 |
| cntn2 | 0.76 |
| LOC100333401 | 0.75 |
| ncanb | 0.75 |
Gene Ontology
Download
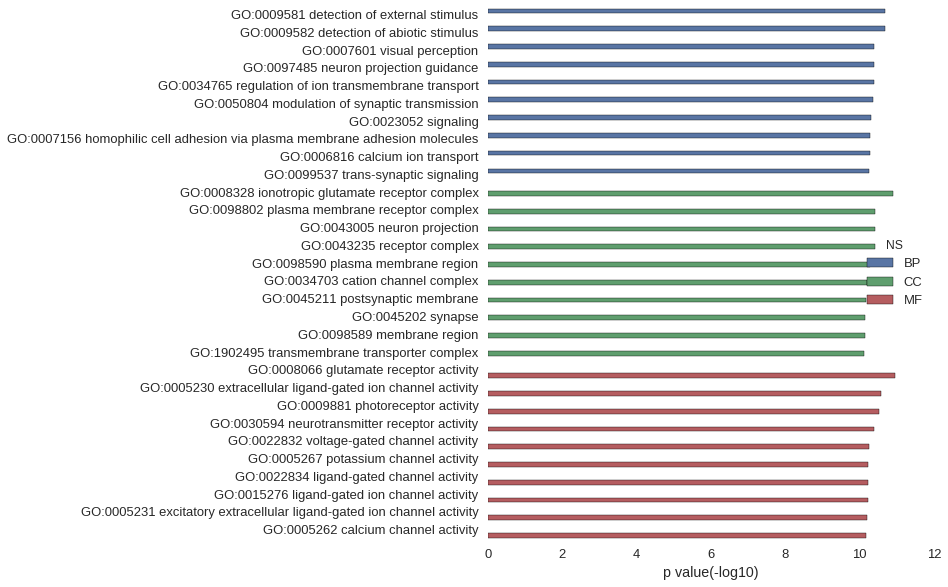
| GO | P value |
|---|---|
| GO:0009581 | 2.13e-11 |
| GO:0009582 | 2.13e-11 |
| GO:0007601 | 4.02e-11 |
| GO:0097485 | 4.08e-11 |
| GO:0034765 | 4.18e-11 |
| GO:0050804 | 4.51e-11 |
| GO:0023052 | 4.88e-11 |
| GO:0007156 | 5.20e-11 |
| GO:0006816 | 5.22e-11 |
| GO:0099537 | 5.52e-11 |
| GO:0008328 | 1.31e-11 |
| GO:0098802 | 3.81e-11 |
| GO:0043005 | 3.81e-11 |
| GO:0043235 | 3.98e-11 |
| GO:0098590 | 5.35e-11 |
| GO:0034703 | 6.25e-11 |
| GO:0045211 | 6.72e-11 |
| GO:0045202 | 7.30e-11 |
| GO:0098589 | 7.33e-11 |
| GO:1902495 | 7.58e-11 |
| GO:0008066 | 1.09e-11 |
| GO:0005230 | 2.60e-11 |
| GO:0009881 | 3.05e-11 |
| GO:0030594 | 4.19e-11 |
| GO:0022832 | 5.77e-11 |
| GO:0005267 | 5.84e-11 |
| GO:0022834 | 6.10e-11 |
| GO:0015276 | 6.10e-11 |
| GO:0005231 | 6.19e-11 |
| GO:0005262 | 6.60e-11 |
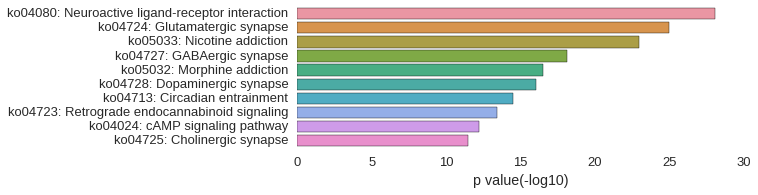
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03008
TCCAGTGCTCCTCATCCGCCAGTCGGTGCAGCAGAAGCGGGTCCGTCAGCATGGAGCTCCGGGCTGCATA
ACGCACACATCCAGGACACACCGCGCTCTCCTACGCACAGATCTGCGGGATTGATTGACTGACTCCCTGC
CTGATTGATTGATTAGCAGCGGATTTCGCGCTCGTGTCGCTCCACCATGGGCTGCGTCACTGGAGATATT
CGCCGACTTTCCGCGCTCCTCATCGGGCTGCAGGTCCTCTACACACACGGTCAGTGCTGCTCAC
TCCAGTGCTCCTCATCCGCCAGTCGGTGCAGCAGAAGCGGGTCCGTCAGCATGGAGCTCCGGGCTGCATA
ACGCACACATCCAGGACACACCGCGCTCTCCTACGCACAGATCTGCGGGATTGATTGACTGACTCCCTGC
CTGATTGATTGATTAGCAGCGGATTTCGCGCTCGTGTCGCTCCACCATGGGCTGCGTCACTGGAGATATT
CGCCGACTTTCCGCGCTCCTCATCGGGCTGCAGGTCCTCTACACACACGGTCAGTGCTGCTCAC