ZFLNCG03592
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700534 | heart | control morpholino | 12.02 |
| SRR1562532 | spleen | normal | 11.72 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 11.32 |
| SRR1647683 | spleen | SVCV treatment | 8.79 |
| SRR1647682 | spleen | normal | 8.23 |
| ERR594417 | retina | normal | 4.44 |
| SRR1647684 | spleen | SVCV treatment | 4.42 |
| SRR1648855 | brain | normal | 4.41 |
| SRR700537 | heart | Gata4 morpholino | 4.05 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 3.99 |
Express in tissues
Correlated coding gene
Download
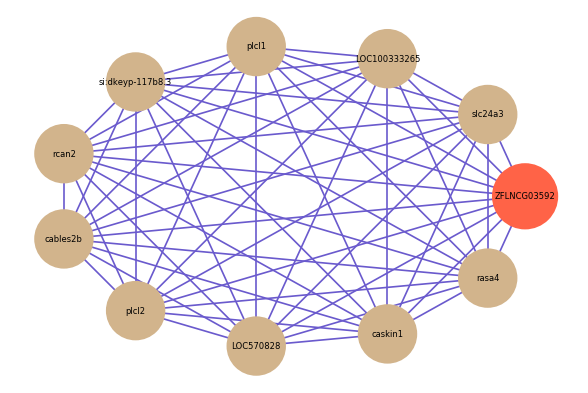
| gene | correlation coefficent |
|---|---|
| slc24a3 | 0.74 |
| LOC100333265 | 0.72 |
| plcl1 | 0.71 |
| si:dkeyp-117b8.3 | 0.71 |
| rcan2 | 0.71 |
| cables2b | 0.71 |
| plcl2 | 0.71 |
| LOC570828 | 0.70 |
| caskin1 | 0.70 |
| rasa4 | 0.70 |
Gene Ontology
Download
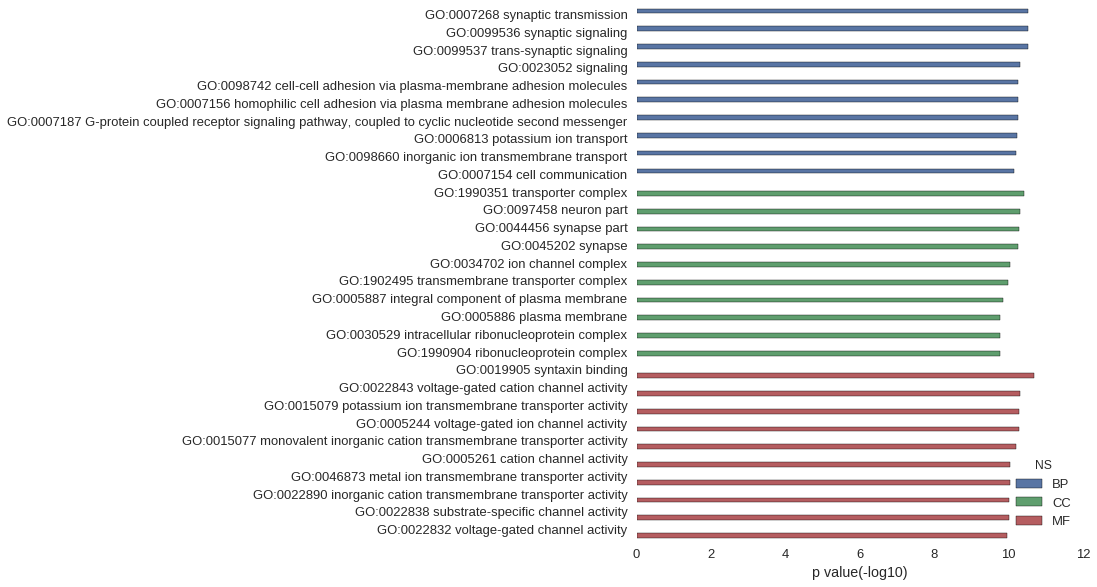
| GO | P value |
|---|---|
| GO:0007268 | 3.04e-11 |
| GO:0099536 | 3.04e-11 |
| GO:0099537 | 3.04e-11 |
| GO:0023052 | 5.02e-11 |
| GO:0098742 | 5.51e-11 |
| GO:0007156 | 5.57e-11 |
| GO:0007187 | 5.58e-11 |
| GO:0006813 | 6.09e-11 |
| GO:0098660 | 6.24e-11 |
| GO:0007154 | 7.26e-11 |
| GO:1990351 | 3.80e-11 |
| GO:0097458 | 5.06e-11 |
| GO:0044456 | 5.34e-11 |
| GO:0045202 | 5.51e-11 |
| GO:0034702 | 8.99e-11 |
| GO:1902495 | 1.02e-10 |
| GO:0005887 | 1.38e-10 |
| GO:0005886 | 1.67e-10 |
| GO:0030529 | 1.72e-10 |
| GO:1990904 | 1.72e-10 |
| GO:0019905 | 2.02e-11 |
| GO:0022843 | 4.99e-11 |
| GO:0015079 | 5.32e-11 |
| GO:0005244 | 5.34e-11 |
| GO:0015077 | 6.45e-11 |
| GO:0005261 | 9.01e-11 |
| GO:0046873 | 9.10e-11 |
| GO:0022890 | 9.85e-11 |
| GO:0022838 | 9.93e-11 |
| GO:0022832 | 1.09e-10 |
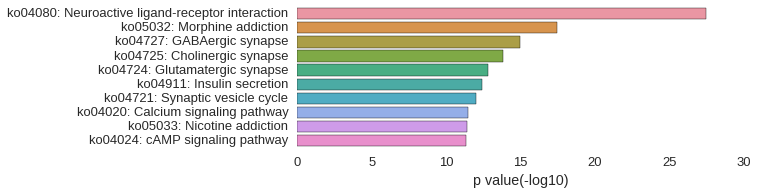
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03592
CTGGCTTTAGTTACCCATTCTAGCCTTCAATATGGCACCAATGTTTATGGCATTGAAGGCAGGCAATCAA
ACATAATCGCTAAAGGTTGTGCCTCTTGTTGCCGAAGAGGTTTTCCATGAACCGCTCTTACTAGCTGGAC
TGTTACACCTAGATGAGCCAGTTAAGGCACTGACGAATGAAGAGCAGACCTGTAGTTATACTGTAAAGGT
GCTCGCTGAGTAATAGAAAGGAGTCAGGGTATTGACATTACAGAGTGCTATAAAGTATCCAGCTGCGGCA
GTGACATTGTAAATCTCTCTCTAGAGGAGACAGTCTTTGCTTTGGGTCAGCCGGTGGGGTTTCGGTCTGG
GTAGTTTGTCACAGTCAGACATTGAATAACAAAGTGGGTCCTAAGGGCTGTGACACATTAGAAAAGCAGA
CAGACATCAGAAACATCATTGGATAATGATATGACAAGAATTCCACAATCAAACTATGGCAGACAAATCT
AATTTAACTTGGAAATAATCAAATAAAGCTTTTTTTAAGAAGCAAAGGAAAGACTCTGCACTGTTTCTTG
GTCATTAATTAATTTAAGCTTAGTTAAATTGAAT
CTGGCTTTAGTTACCCATTCTAGCCTTCAATATGGCACCAATGTTTATGGCATTGAAGGCAGGCAATCAA
ACATAATCGCTAAAGGTTGTGCCTCTTGTTGCCGAAGAGGTTTTCCATGAACCGCTCTTACTAGCTGGAC
TGTTACACCTAGATGAGCCAGTTAAGGCACTGACGAATGAAGAGCAGACCTGTAGTTATACTGTAAAGGT
GCTCGCTGAGTAATAGAAAGGAGTCAGGGTATTGACATTACAGAGTGCTATAAAGTATCCAGCTGCGGCA
GTGACATTGTAAATCTCTCTCTAGAGGAGACAGTCTTTGCTTTGGGTCAGCCGGTGGGGTTTCGGTCTGG
GTAGTTTGTCACAGTCAGACATTGAATAACAAAGTGGGTCCTAAGGGCTGTGACACATTAGAAAAGCAGA
CAGACATCAGAAACATCATTGGATAATGATATGACAAGAATTCCACAATCAAACTATGGCAGACAAATCT
AATTTAACTTGGAAATAATCAAATAAAGCTTTTTTTAAGAAGCAAAGGAAAGACTCTGCACTGTTTCTTG
GTCATTAATTAATTTAAGCTTAGTTAAATTGAAT