ZFLNCG03758
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941749 | anterior pectoral fin | normal | 9.09 |
| SRR516128 | skin | male and 5 month | 8.38 |
| SRR957180 | heart | 7 days after heart tip amputation | 8.19 |
| SRR516130 | skin | male and 5 month | 7.59 |
| SRR516129 | skin | male and 5 month | 7.49 |
| SRR891504 | liver | normal | 7.39 |
| SRR957181 | heart | normal | 7.38 |
| SRR527834 | head | normal | 7.38 |
| SRR941753 | posterior pectoral fin | normal | 7.34 |
| SRR1562531 | muscle | normal | 7.02 |
Express in tissues
Correlated coding gene
Download
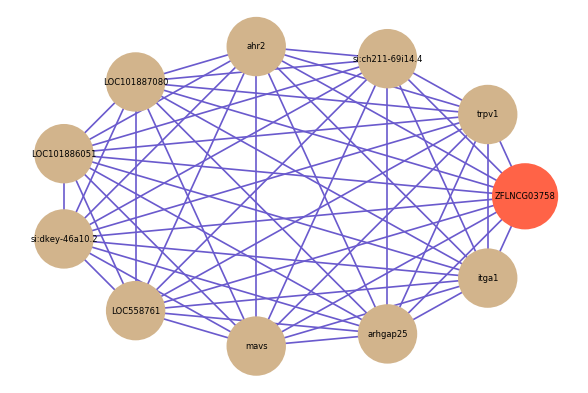
| gene | correlation coefficent |
|---|---|
| trpv1 | 0.68 |
| si:ch211-69i14.4 | 0.67 |
| ahr2 | 0.67 |
| LOC101887080 | 0.66 |
| LOC101886051 | 0.66 |
| si:dkey-46a10.2 | 0.65 |
| LOC558761 | 0.65 |
| mavs | 0.64 |
| arhgap25 | 0.64 |
| itga1 | 0.64 |
Gene Ontology
Download
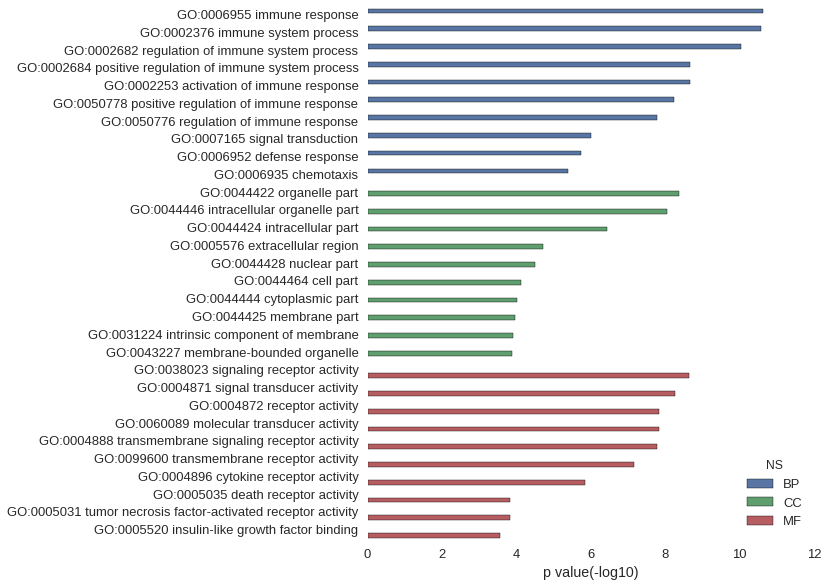
| GO | P value |
|---|---|
| GO:0006955 | 2.28e-11 |
| GO:0002376 | 2.72e-11 |
| GO:0002682 | 9.41e-11 |
| GO:0002684 | 2.11e-09 |
| GO:0002253 | 2.13e-09 |
| GO:0050778 | 5.81e-09 |
| GO:0050776 | 1.61e-08 |
| GO:0007165 | 9.70e-07 |
| GO:0006952 | 1.83e-06 |
| GO:0006935 | 4.07e-06 |
| GO:0044422 | 4.25e-09 |
| GO:0044446 | 8.78e-09 |
| GO:0044424 | 3.76e-07 |
| GO:0005576 | 1.92e-05 |
| GO:0044428 | 3.08e-05 |
| GO:0044464 | 7.29e-05 |
| GO:0044444 | 9.33e-05 |
| GO:0044425 | 1.12e-04 |
| GO:0031224 | 1.25e-04 |
| GO:0043227 | 1.35e-04 |
| GO:0038023 | 2.30e-09 |
| GO:0004871 | 5.34e-09 |
| GO:0004872 | 1.46e-08 |
| GO:0060089 | 1.46e-08 |
| GO:0004888 | 1.63e-08 |
| GO:0099600 | 6.84e-08 |
| GO:0004896 | 1.46e-06 |
| GO:0005035 | 1.45e-04 |
| GO:0005031 | 1.45e-04 |
| GO:0005520 | 2.80e-04 |
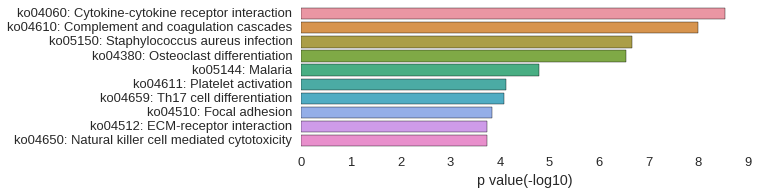
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03758
TGTGTTGGGTGTGTTTGTATGGGATTTCTGAATCAGCTTTCTGAAATGCTCGTCTGAGTTAGCTAAATTC
AGTAGGACTGAGGGCGACAGAAGCGAACTGACTCTTCATTCAACCTGGCTGTAACTAAATATAGCCACAC
ACTCTCAAACTTGCATGAGTGCTGACGACAAGTGGTGGTCGATGGTAGGAATCCGGAGCGGGTGTGTTTG
CTGCTTTTGTGGTTGTGTGTGTTCAAAGATTGTGTGTGTGCGTGTTAACCATTAATCAGATTCTCTCCAT
GTTTCTCTAGGATTCTGGGAATGTTCTTTAAACCAACAACACAGGACATTTTCTGCCCCTGGCAGTCTTC
TAGTTCAGGGCAGGACATCGATCTGTATAGAGGGCACCTGCTCTGATGTATTACCGGGAGAGGATCAAAC
TTGCAGCCCAACTGGGAATCTAATCCCTCCATTGACCGACAACAGCCCTAATGCCTCCAAAGGTGAAAGT
AGTAGTGCCCTGTCTTTTATAAACCCAACTTTCCTCAAGACACAGCAAGATCTCAGTCAAAATGCAGCAA
CTGTGATCACTGACG
TGTGTTGGGTGTGTTTGTATGGGATTTCTGAATCAGCTTTCTGAAATGCTCGTCTGAGTTAGCTAAATTC
AGTAGGACTGAGGGCGACAGAAGCGAACTGACTCTTCATTCAACCTGGCTGTAACTAAATATAGCCACAC
ACTCTCAAACTTGCATGAGTGCTGACGACAAGTGGTGGTCGATGGTAGGAATCCGGAGCGGGTGTGTTTG
CTGCTTTTGTGGTTGTGTGTGTTCAAAGATTGTGTGTGTGCGTGTTAACCATTAATCAGATTCTCTCCAT
GTTTCTCTAGGATTCTGGGAATGTTCTTTAAACCAACAACACAGGACATTTTCTGCCCCTGGCAGTCTTC
TAGTTCAGGGCAGGACATCGATCTGTATAGAGGGCACCTGCTCTGATGTATTACCGGGAGAGGATCAAAC
TTGCAGCCCAACTGGGAATCTAATCCCTCCATTGACCGACAACAGCCCTAATGCCTCCAAAGGTGAAAGT
AGTAGTGCCCTGTCTTTTATAAACCCAACTTTCCTCAAGACACAGCAAGATCTCAGTCAAAATGCAGCAA
CTGTGATCACTGACG