ZFLNCG03788
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562529 | intestine and pancreas | normal | 351.59 |
| SRR891512 | blood | normal | 49.34 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 37.83 |
| SRR516121 | skin | male and 3.5 year | 18.88 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 18.14 |
| SRR516123 | skin | male and 3.5 year | 15.53 |
| SRR1562528 | eye | normal | 15.29 |
| SRR941749 | anterior pectoral fin | normal | 13.36 |
| SRR941753 | posterior pectoral fin | normal | 12.93 |
| SRR516124 | skin | male and 3.5 year | 12.54 |
Express in tissues
Correlated coding gene
Download
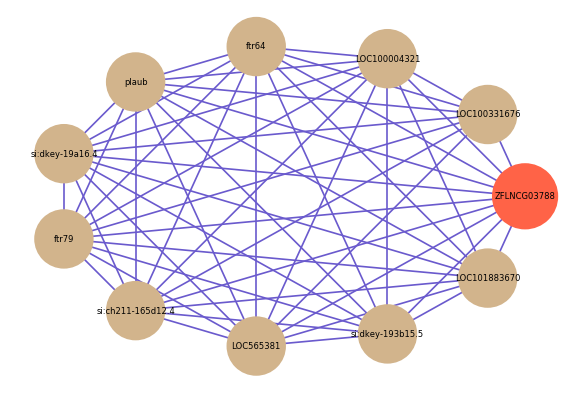
| gene | correlation coefficent |
|---|---|
| LOC100331676 | 0.65 |
| LOC100004321 | 0.63 |
| ftr64 | 0.63 |
| plaub | 0.63 |
| si:dkey-19a16.4 | 0.63 |
| si:dkey-193b15.5 | 0.63 |
| ftr79 | 0.62 |
| si:ch211-165d12.4 | 0.62 |
| LOC101883670 | 0.62 |
| LOC565381 | 0.61 |
Gene Ontology
Download
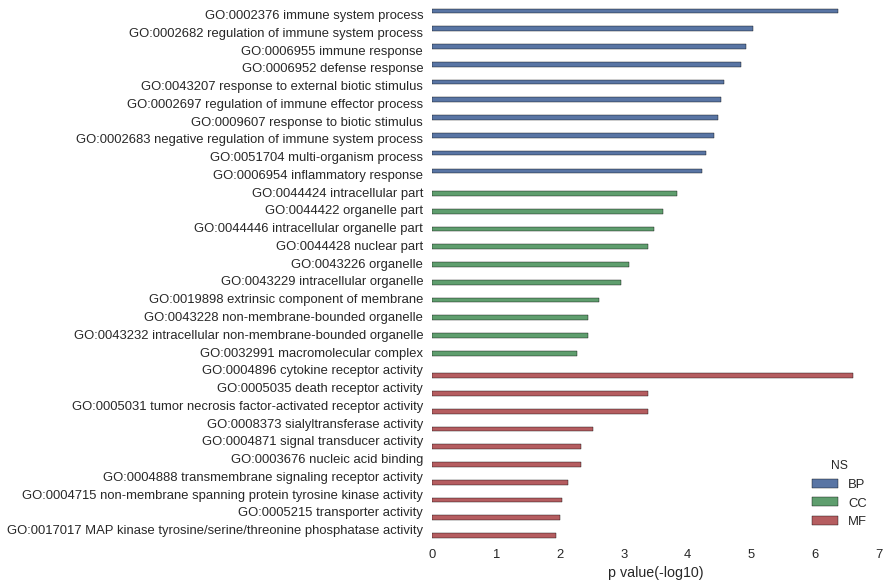
| GO | P value |
|---|---|
| GO:0002376 | 4.30e-07 |
| GO:0002682 | 9.42e-06 |
| GO:0006955 | 1.20e-05 |
| GO:0006952 | 1.42e-05 |
| GO:0043207 | 2.65e-05 |
| GO:0002697 | 2.97e-05 |
| GO:0009607 | 3.35e-05 |
| GO:0002683 | 3.85e-05 |
| GO:0051704 | 5.10e-05 |
| GO:0006954 | 5.80e-05 |
| GO:0044424 | 1.48e-04 |
| GO:0044422 | 2.41e-04 |
| GO:0044446 | 3.36e-04 |
| GO:0044428 | 4.16e-04 |
| GO:0043226 | 8.30e-04 |
| GO:0043229 | 1.10e-03 |
| GO:0019898 | 2.47e-03 |
| GO:0043228 | 3.63e-03 |
| GO:0043232 | 3.63e-03 |
| GO:0032991 | 5.34e-03 |
| GO:0004896 | 2.49e-07 |
| GO:0005035 | 4.15e-04 |
| GO:0005031 | 4.15e-04 |
| GO:0008373 | 3.01e-03 |
| GO:0004871 | 4.70e-03 |
| GO:0003676 | 4.71e-03 |
| GO:0004888 | 7.36e-03 |
| GO:0004715 | 9.18e-03 |
| GO:0005215 | 9.85e-03 |
| GO:0017017 | 1.13e-02 |
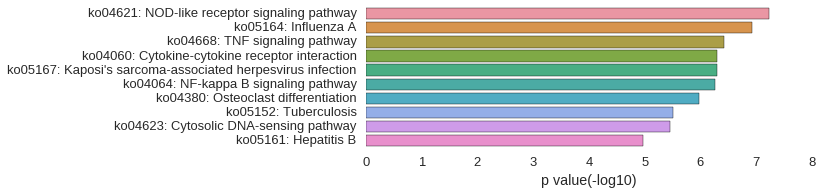
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG03788
ACCTCTTTCAGCAGAATCCCATAGGGACGAATGGACTTTTCCACAATCTCCTTCTCTTGCTGGGCAGAAG
TGCCATCAGGAACTGGAATCAGCCAATAGTCTTTTCTGTCACATATCTTCCTCATCTTTGGGTTGATCCT
GTTCTGCATTATGACTGTATCTCTTCCCGGTCTTCTTGGATGTGAACTCTCTGCTGTATCCAGAAGCCTG
ATCGATGTCATATGTCGAGTAGATTCCTCTTCCGTACAGATCTCCAGATCCTGCTTTATAATGCGTTTTG
ACGATTCCTCTTGCACCTTCAGCAGAGGTTCCGTGATAAGACACAGGCCACTCGCCGGCCACCGAGTAAC
TCCTCAATCCATCCGTCCCCAGCCAAGAGTTTCCATCTGGATACTTGTTCAGGACTTTCAGAGCAATGCG
GTACCATCCACATGGCCGAATATACGGCTCATTGCCTCTCATGCAAACTGACTTGTCTCTTGAATGTGTA
AAGTCATAATCAAAGCGCGGTGCGAAGAATTCATCCATGTCTACAAACATCTCTGTGTGTACATCAGAGT
TGAATCCTGCTGCTGACGCTGAATC
ACCTCTTTCAGCAGAATCCCATAGGGACGAATGGACTTTTCCACAATCTCCTTCTCTTGCTGGGCAGAAG
TGCCATCAGGAACTGGAATCAGCCAATAGTCTTTTCTGTCACATATCTTCCTCATCTTTGGGTTGATCCT
GTTCTGCATTATGACTGTATCTCTTCCCGGTCTTCTTGGATGTGAACTCTCTGCTGTATCCAGAAGCCTG
ATCGATGTCATATGTCGAGTAGATTCCTCTTCCGTACAGATCTCCAGATCCTGCTTTATAATGCGTTTTG
ACGATTCCTCTTGCACCTTCAGCAGAGGTTCCGTGATAAGACACAGGCCACTCGCCGGCCACCGAGTAAC
TCCTCAATCCATCCGTCCCCAGCCAAGAGTTTCCATCTGGATACTTGTTCAGGACTTTCAGAGCAATGCG
GTACCATCCACATGGCCGAATATACGGCTCATTGCCTCTCATGCAAACTGACTTGTCTCTTGAATGTGTA
AAGTCATAATCAAAGCGCGGTGCGAAGAATTCATCCATGTCTACAAACATCTCTGTGTGTACATCAGAGT
TGAATCCTGCTGCTGACGCTGAATC