ZFLNCG04191
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 327.05 |
| SRR1648854 | brain | normal | 205.62 |
| SRR519727 | 6 dpf | FETOH treatment | 191.40 |
| SRR592703 | pineal gland | normal | 187.49 |
| SRR1028004 | head | normal | 181.87 |
| SRR519752 | 7 dpf | VD3 treatment | 157.75 |
| SRR519747 | 6 dpf | VD3 treatment | 151.73 |
| SRR519722 | 4 dpf | FETOH treatment | 150.50 |
| SRR519732 | 7 dpf | FETOH treatment | 138.59 |
| SRR372802 | 5 dpf | normal | 137.92 |
Express in tissues
Correlated coding gene
Download
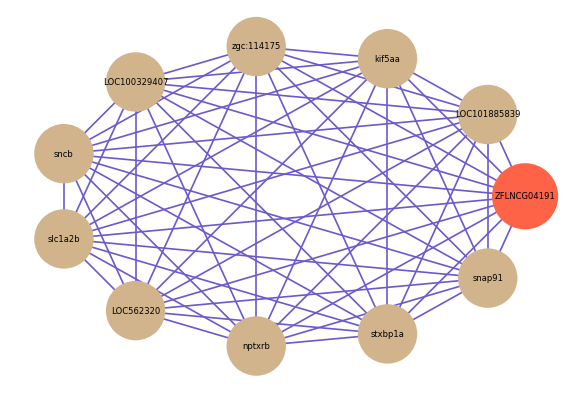
| gene | correlation coefficent |
|---|---|
| LOC101885839 | 0.87 |
| kif5aa | 0.87 |
| zgc:114175 | 0.87 |
| LOC100329407 | 0.87 |
| sncb | 0.87 |
| slc1a2b | 0.87 |
| LOC562320 | 0.86 |
| nptxrb | 0.86 |
| stxbp1a | 0.86 |
| snap91 | 0.86 |
Gene Ontology
Download
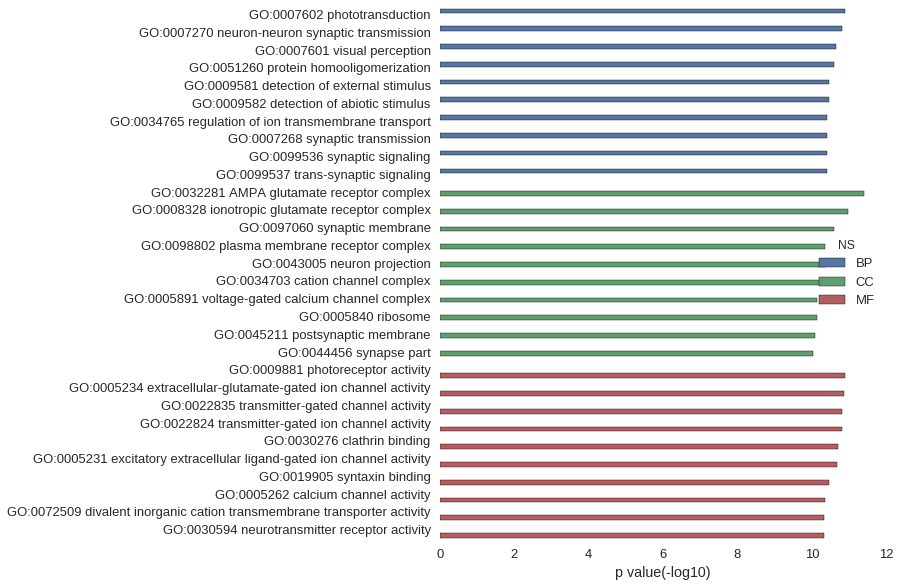
| GO | P value |
|---|---|
| GO:0007602 | 1.31e-11 |
| GO:0007270 | 1.57e-11 |
| GO:0007601 | 2.30e-11 |
| GO:0051260 | 2.61e-11 |
| GO:0009581 | 3.54e-11 |
| GO:0009582 | 3.54e-11 |
| GO:0034765 | 3.91e-11 |
| GO:0007268 | 3.95e-11 |
| GO:0099536 | 3.95e-11 |
| GO:0099537 | 3.95e-11 |
| GO:0032281 | 3.90e-12 |
| GO:0008328 | 1.05e-11 |
| GO:0097060 | 2.55e-11 |
| GO:0098802 | 4.52e-11 |
| GO:0043005 | 4.52e-11 |
| GO:0034703 | 4.68e-11 |
| GO:0005891 | 7.23e-11 |
| GO:0005840 | 7.33e-11 |
| GO:0045211 | 8.37e-11 |
| GO:0044456 | 9.19e-11 |
| GO:0009881 | 1.31e-11 |
| GO:0005234 | 1.40e-11 |
| GO:0022835 | 1.51e-11 |
| GO:0022824 | 1.51e-11 |
| GO:0030276 | 1.96e-11 |
| GO:0005231 | 2.04e-11 |
| GO:0019905 | 3.54e-11 |
| GO:0005262 | 4.36e-11 |
| GO:0072509 | 4.58e-11 |
| GO:0030594 | 4.61e-11 |
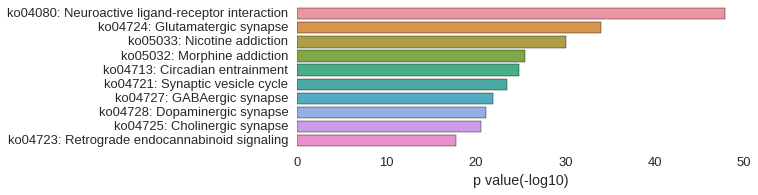
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04191
GTGTGTGAAAACTGCCAGTTTTTATAAACAATAATGATAATTTTTGTAATTAGTTAACTTTTTTTAAGAA
AATGTTTATACTTGGCTGTGATGGCTATGATCTCAACACCTTAGAACATAGATATTAACATTTGATGTTG
CCTATGCTGTTGTTGTCTATTGGCGTAGATATATACACTAGATATCGCATTTGGATCCTGATCATGC
GTGTGTGAAAACTGCCAGTTTTTATAAACAATAATGATAATTTTTGTAATTAGTTAACTTTTTTTAAGAA
AATGTTTATACTTGGCTGTGATGGCTATGATCTCAACACCTTAGAACATAGATATTAACATTTGATGTTG
CCTATGCTGTTGTTGTCTATTGGCGTAGATATATACACTAGATATCGCATTTGGATCCTGATCATGC