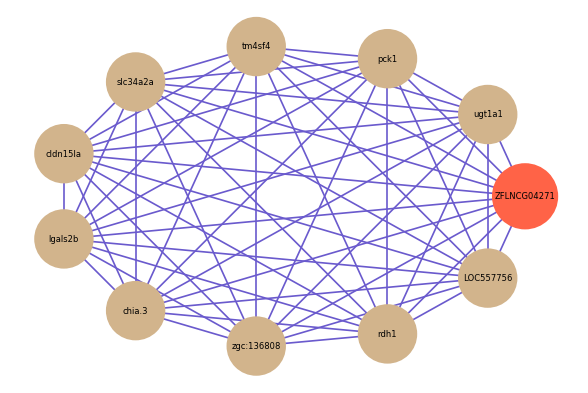
ZFLNCG04271
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035239 | liver | transgenic mCherry control | 304.18 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 125.32 |
| SRR1562529 | intestine and pancreas | normal | 123.98 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 103.93 |
| SRR527833 | 5 dpf | normal | 78.90 |
| SRR1035237 | liver | transgenic UHRF1 | 66.80 |
| SRR1188148 | embryo | Control PBS 0.5 hpi | 38.90 |
| SRR1291414 | 5 dpi | normal | 38.00 |
| SRR1562530 | liver | normal | 37.89 |
| SRR1188156 | embryo | Control PBS 4 hpi | 32.41 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
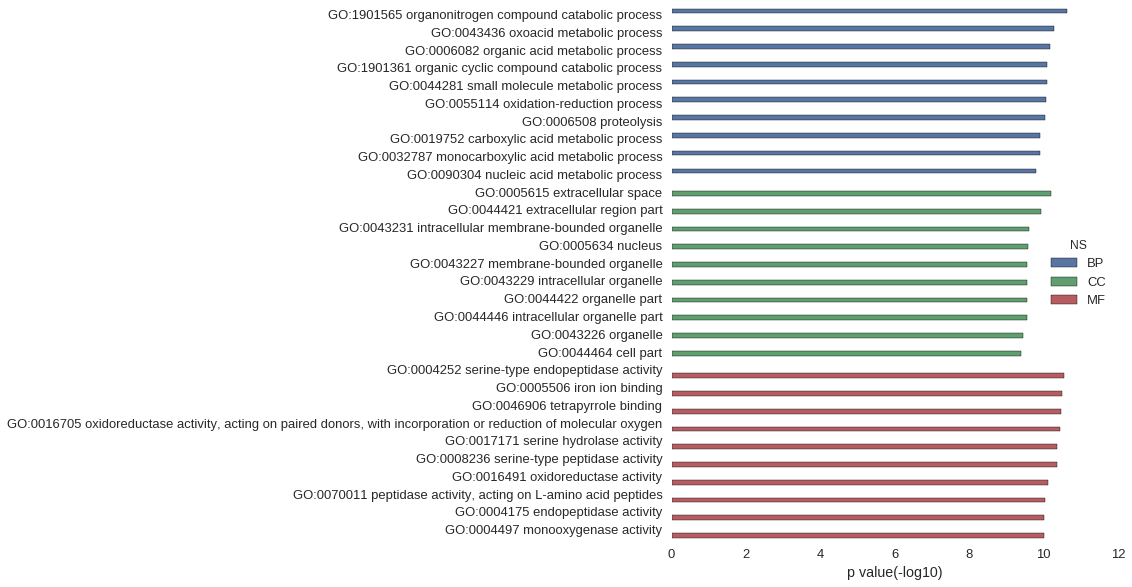
| GO | P value |
|---|---|
| GO:1901565 | 2.35e-11 |
| GO:0043436 | 5.24e-11 |
| GO:0006082 | 6.88e-11 |
| GO:1901361 | 8.01e-11 |
| GO:0044281 | 8.05e-11 |
| GO:0055114 | 8.69e-11 |
| GO:0006508 | 9.20e-11 |
| GO:0019752 | 1.21e-10 |
| GO:0032787 | 1.22e-10 |
| GO:0090304 | 1.58e-10 |
| GO:0005615 | 6.38e-11 |
| GO:0044421 | 1.14e-10 |
| GO:0043231 | 2.50e-10 |
| GO:0005634 | 2.69e-10 |
| GO:0043227 | 2.77e-10 |
| GO:0043229 | 2.78e-10 |
| GO:0044422 | 2.79e-10 |
| GO:0044446 | 2.86e-10 |
| GO:0043226 | 3.59e-10 |
| GO:0044464 | 4.08e-10 |
| GO:0004252 | 2.82e-11 |
| GO:0005506 | 3.11e-11 |
| GO:0046906 | 3.45e-11 |
| GO:0016705 | 3.69e-11 |
| GO:0017171 | 4.25e-11 |
| GO:0008236 | 4.25e-11 |
| GO:0016491 | 7.81e-11 |
| GO:0070011 | 8.94e-11 |
| GO:0004175 | 9.68e-11 |
| GO:0004497 | 9.71e-11 |
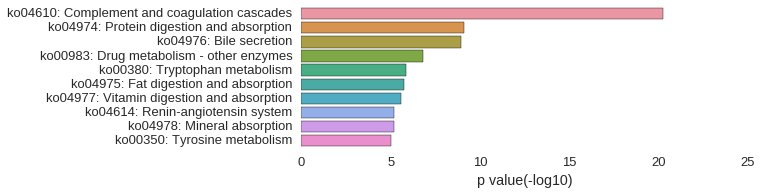
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04271
ACTCACTGCTGGGGAAGAAGTGCTTCGCTCTGCGCATCGCATCACGCATCGCTAAAGAGGAGGGCTGGCT
GGCAGAACACATGCTGGTGAGTGGACGAACACACACACACACACACACACACATGCAGAAGCAGAACACA
TGCTGCTCAGCGGAGAAAACACACACACAACTGCAGAGTCATTCTGACTGGGCAGACCTCACTAAACACT
GACAGATGGAGCGTTAACACACACATGCATGTACACACACACACACATATGCACACACACATGTTTACAC
TTCTTTCAATGCAGTTTGTACAGTATCACAGTTATTATTTTAATATGGGGTTATCAGAGAATAACGAACC
TTCAGATGTTGTGATGGACCAATCAGAATCTAGTATTATATTGATTGTATGAAGCAGTAATTAATGTAAA
CAGCTCTACACAAATGAACTTGAGTTAAAGTGTGCTGAATTTCCCCTGCAGATTCTGGGCATCACCAACC
CGGCGGGACAGAAGAAGTATTTCGCAGCGGCGTTCCCCAGCGCCTGCGGGAAGACCAACCTGGCCATGCT
GAAGCCGTCGCTGCCCGGCTGGAAGGTGGAGTGTGTGGGAGACGACATCGCATGGATGAAGTTCGACAAA
GAAGGTACAGAGACAAACACACACACACACACACACACACAGCATTCTGGATTACTGGATTTAGCTCACG
TTCTGCTTATCATAACTCAATCTTCATTCTACAGCTCTTCTAGTAAATGAAAGTCTTTTAGATCTGTACA
TATCAAACGCTTGTTTTTGGGCCAGTAAGAATATCCTGAGCTCAAACTGTGGATCTCCAGGACTGAGAGT
TTATTTGATGGTTCTTTCTTCATGTTTCCTCAGGGAATCTGAGAGCCATCAACCCAGAGAACGGCTTCTT
CGGTGTGGCTCCGGGAACCTCCAGCAAAACCAACCCCAACGCCATGAGCACCATCAGCTGCAACACACTC
TTCACCAACGTGGCGGAGAGCAGTGACGGAGGGGTGTTCTGGGAGGGCATGGATGAGGATCTGCCTGAAG
GAGTGACGCTGACGTCCTGGAAGAACCAGCCCTGGACACCAGAGGACGGTGAGTGACATCTGAAATGATC
AGCTTCATCTTTAACTGTTCACTCTTCCTCAAGAGGTCGTAATCCCTCAGGAGTTTCATTCCTCTGCTGA
ACATAAAGGAAGATATTTGGAAGAATGCTGAAAACCCAATGTAGGAAAAGCAAATAGAGTCAATGAATAC
AGGTTTGGAGATTTCTTCAAAATATCTTCTTCTGTGTCTAATAGAAGAAAGAAACTTGGTAAAGGATGAG
G
ACTCACTGCTGGGGAAGAAGTGCTTCGCTCTGCGCATCGCATCACGCATCGCTAAAGAGGAGGGCTGGCT
GGCAGAACACATGCTGGTGAGTGGACGAACACACACACACACACACACACACATGCAGAAGCAGAACACA
TGCTGCTCAGCGGAGAAAACACACACACAACTGCAGAGTCATTCTGACTGGGCAGACCTCACTAAACACT
GACAGATGGAGCGTTAACACACACATGCATGTACACACACACACACATATGCACACACACATGTTTACAC
TTCTTTCAATGCAGTTTGTACAGTATCACAGTTATTATTTTAATATGGGGTTATCAGAGAATAACGAACC
TTCAGATGTTGTGATGGACCAATCAGAATCTAGTATTATATTGATTGTATGAAGCAGTAATTAATGTAAA
CAGCTCTACACAAATGAACTTGAGTTAAAGTGTGCTGAATTTCCCCTGCAGATTCTGGGCATCACCAACC
CGGCGGGACAGAAGAAGTATTTCGCAGCGGCGTTCCCCAGCGCCTGCGGGAAGACCAACCTGGCCATGCT
GAAGCCGTCGCTGCCCGGCTGGAAGGTGGAGTGTGTGGGAGACGACATCGCATGGATGAAGTTCGACAAA
GAAGGTACAGAGACAAACACACACACACACACACACACACAGCATTCTGGATTACTGGATTTAGCTCACG
TTCTGCTTATCATAACTCAATCTTCATTCTACAGCTCTTCTAGTAAATGAAAGTCTTTTAGATCTGTACA
TATCAAACGCTTGTTTTTGGGCCAGTAAGAATATCCTGAGCTCAAACTGTGGATCTCCAGGACTGAGAGT
TTATTTGATGGTTCTTTCTTCATGTTTCCTCAGGGAATCTGAGAGCCATCAACCCAGAGAACGGCTTCTT
CGGTGTGGCTCCGGGAACCTCCAGCAAAACCAACCCCAACGCCATGAGCACCATCAGCTGCAACACACTC
TTCACCAACGTGGCGGAGAGCAGTGACGGAGGGGTGTTCTGGGAGGGCATGGATGAGGATCTGCCTGAAG
GAGTGACGCTGACGTCCTGGAAGAACCAGCCCTGGACACCAGAGGACGGTGAGTGACATCTGAAATGATC
AGCTTCATCTTTAACTGTTCACTCTTCCTCAAGAGGTCGTAATCCCTCAGGAGTTTCATTCCTCTGCTGA
ACATAAAGGAAGATATTTGGAAGAATGCTGAAAACCCAATGTAGGAAAAGCAAATAGAGTCAATGAATAC
AGGTTTGGAGATTTCTTCAAAATATCTTCTTCTGTGTCTAATAGAAGAAAGAAACTTGGTAAAGGATGAG
G