ZFLNCG04495
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR748488 | sperm | normal | 214.76 |
| SRR527835 | tail | normal | 93.45 |
| SRR658547 | 6 somite | Gata5/6 morphant | 36.82 |
| SRR1647679 | head kidney | normal | 35.26 |
| SRR658539 | bud | Gata5/6 morphant | 33.64 |
| SRR658533 | bud | normal | 31.64 |
| SRR658535 | bud | Gata5 morphant | 25.16 |
| SRR1647681 | head kidney | SVCV treatment | 24.49 |
| SRR372796 | bud | normal | 23.76 |
| SRR1004785 | larvae | normal | 19.66 |
Express in tissues
Correlated coding gene
Download
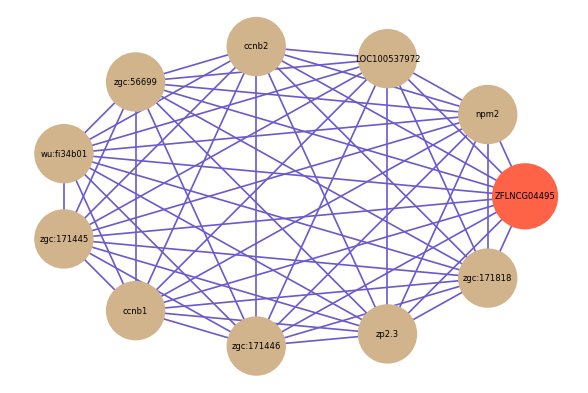
| gene | correlation coefficent |
|---|---|
| npm2 | 0.89 |
| LOC100537972 | 0.86 |
| ccnb2 | 0.86 |
| zgc:56699 | 0.85 |
| wu:fi34b01 | 0.85 |
| zgc:171445 | 0.83 |
| ccnb1 | 0.83 |
| zgc:171446 | 0.83 |
| zp2.3 | 0.83 |
| zgc:171818 | 0.83 |
Gene Ontology
Download
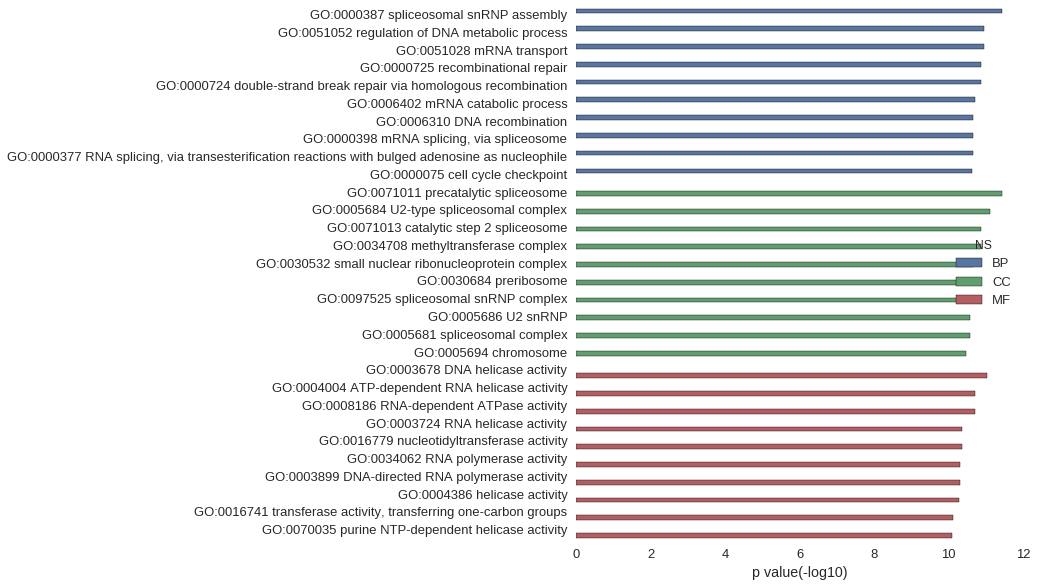
| GO | P value |
|---|---|
| GO:0000387 | 3.62e-12 |
| GO:0051052 | 1.09e-11 |
| GO:0051028 | 1.09e-11 |
| GO:0000725 | 1.35e-11 |
| GO:0000724 | 1.35e-11 |
| GO:0006402 | 1.89e-11 |
| GO:0006310 | 2.12e-11 |
| GO:0000398 | 2.12e-11 |
| GO:0000377 | 2.12e-11 |
| GO:0000075 | 2.36e-11 |
| GO:0071011 | 3.62e-12 |
| GO:0005684 | 7.60e-12 |
| GO:0071013 | 1.35e-11 |
| GO:0034708 | 1.35e-11 |
| GO:0030532 | 2.17e-11 |
| GO:0030684 | 2.17e-11 |
| GO:0097525 | 2.17e-11 |
| GO:0005686 | 2.66e-11 |
| GO:0005681 | 2.68e-11 |
| GO:0005694 | 3.45e-11 |
| GO:0003678 | 9.11e-12 |
| GO:0004004 | 1.89e-11 |
| GO:0008186 | 1.89e-11 |
| GO:0003724 | 4.25e-11 |
| GO:0016779 | 4.39e-11 |
| GO:0034062 | 4.93e-11 |
| GO:0003899 | 4.93e-11 |
| GO:0004386 | 5.32e-11 |
| GO:0016741 | 7.51e-11 |
| GO:0070035 | 7.98e-11 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04495
GCACGCGCTCACACACAGTCAGTGCAGAATTTCTCTCAGAAGTTGTCGGCGGTGCCCTCCATCATGTTTG
GATTTGTACGAGCGCTTAAAAGAACGAGATGTACACCTGGACAACGTGTCTTTCCCTTTAGCCACCGTTT
CTACATTAACTCCAAATCAGCTAAAAGCAGTGTCGGCGTTGTGGTAAGTTTAACTAAGCGAGCTTTTCCT
GTCCTAAACATCTCAGAGTTACAGGCTAATAATTGACGTTTTCGCTGATTTATTAAACGCATTAATAGTG
TGCAATCATTATTTACTTATTAACAACATGATTTGCTTTTAACCACCAATCTGCATACATTTTGAGTAGT
TTTATCAGTCTCTATTGTTTAAACTGGTAATTCTCCAGTATAACTAGTAAGTTAAACTTCCTGATTTTAA
TTAGTAGCTTGAATTGTTCTCTTTCACTGTAACATTAATTCATATTCAACTGATTGGTTTTAAAGATTAC
ATTCAAACATAACAATTCATGGCATCCTTTTTAGTGCAAATGCACAGGTTTATCAGATATACTGTGCCAG
TTTCAGTTTACATCAGCACATTATTATACACTAATGCTCAATGTTGCGTTTATTCCCACTTTTAGGAGAG
CGTATTTGTGATGACCTTCATCTCTCTTGCTGTTCTTGGTCCGGCAGCATGGTTGCTGTCCAAATCATCT
GATACCCACAAAAAACAGCAGTGAGGAAGAATGACGATGCCTTCTGCCAGAGGACCAGACATCTGTGTAT
TCTAGCAGTTTCAGTTGTACACTAACCTACCATCAGTGTGTGTCAATGTCAAATATGCATTTAAATATGG
TGATCAAATACAACCTTTTCAATACCC
GCACGCGCTCACACACAGTCAGTGCAGAATTTCTCTCAGAAGTTGTCGGCGGTGCCCTCCATCATGTTTG
GATTTGTACGAGCGCTTAAAAGAACGAGATGTACACCTGGACAACGTGTCTTTCCCTTTAGCCACCGTTT
CTACATTAACTCCAAATCAGCTAAAAGCAGTGTCGGCGTTGTGGTAAGTTTAACTAAGCGAGCTTTTCCT
GTCCTAAACATCTCAGAGTTACAGGCTAATAATTGACGTTTTCGCTGATTTATTAAACGCATTAATAGTG
TGCAATCATTATTTACTTATTAACAACATGATTTGCTTTTAACCACCAATCTGCATACATTTTGAGTAGT
TTTATCAGTCTCTATTGTTTAAACTGGTAATTCTCCAGTATAACTAGTAAGTTAAACTTCCTGATTTTAA
TTAGTAGCTTGAATTGTTCTCTTTCACTGTAACATTAATTCATATTCAACTGATTGGTTTTAAAGATTAC
ATTCAAACATAACAATTCATGGCATCCTTTTTAGTGCAAATGCACAGGTTTATCAGATATACTGTGCCAG
TTTCAGTTTACATCAGCACATTATTATACACTAATGCTCAATGTTGCGTTTATTCCCACTTTTAGGAGAG
CGTATTTGTGATGACCTTCATCTCTCTTGCTGTTCTTGGTCCGGCAGCATGGTTGCTGTCCAAATCATCT
GATACCCACAAAAAACAGCAGTGAGGAAGAATGACGATGCCTTCTGCCAGAGGACCAGACATCTGTGTAT
TCTAGCAGTTTCAGTTGTACACTAACCTACCATCAGTGTGTGTCAATGTCAAATATGCATTTAAATATGG
TGATCAAATACAACCTTTTCAATACCC