ZFLNCG04733
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800043 | sphere stage | normal | 29.48 |
| SRR1021215 | sphere stage | Mzeomesa | 25.77 |
| SRR514030 | retina | id2a morpholino | 23.39 |
| SRR1021213 | sphere stage | normal | 17.59 |
| SRR800049 | sphere stage | control treatment | 16.70 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 14.33 |
| SRR800046 | sphere stage | 5azaCyD treatment | 11.62 |
| SRR1569495 | embryo | normal | 10.46 |
| SRR372791 | dome | normal | 10.20 |
| SRR065197 | 3 dpf | U1C knockout | 9.35 |
Express in tissues
Correlated coding gene
Download
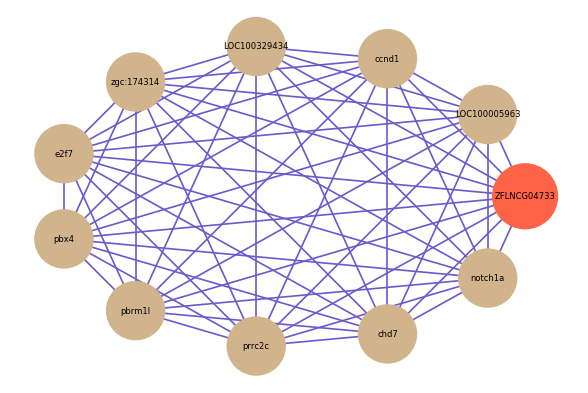
| gene | correlation coefficent |
|---|---|
| LOC100005963 | 0.76 |
| ccnd1 | 0.75 |
| LOC100329434 | 0.74 |
| zgc:174314 | 0.73 |
| e2f7 | 0.72 |
| pbx4 | 0.72 |
| pbrm1l | 0.72 |
| prrc2c | 0.72 |
| chd7 | 0.72 |
| notch1a | 0.72 |
Gene Ontology
Download
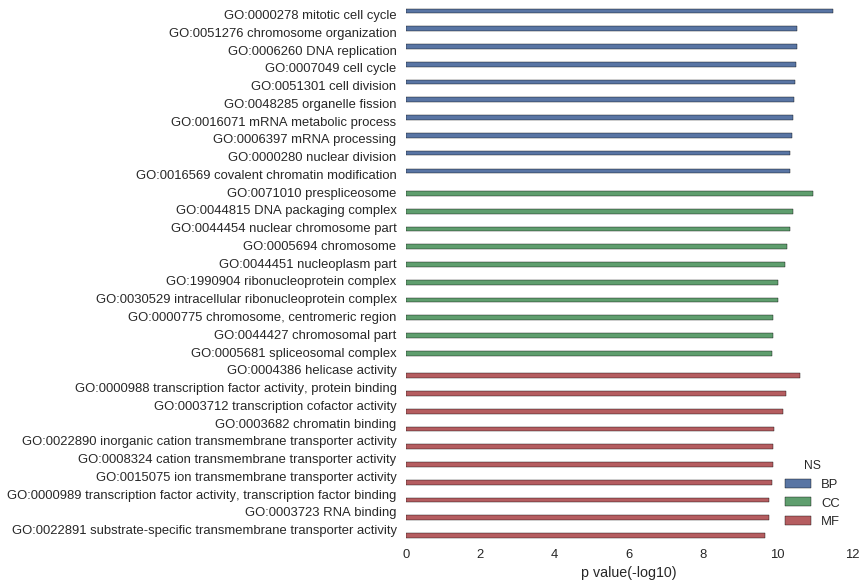
| GO | P value |
|---|---|
| GO:0000278 | 3.29e-12 |
| GO:0051276 | 2.93e-11 |
| GO:0006260 | 2.93e-11 |
| GO:0007049 | 3.29e-11 |
| GO:0051301 | 3.52e-11 |
| GO:0048285 | 3.58e-11 |
| GO:0016071 | 3.97e-11 |
| GO:0006397 | 4.06e-11 |
| GO:0000280 | 4.54e-11 |
| GO:0016569 | 4.65e-11 |
| GO:0071010 | 1.10e-11 |
| GO:0044815 | 3.96e-11 |
| GO:0044454 | 4.65e-11 |
| GO:0005694 | 5.73e-11 |
| GO:0044451 | 6.25e-11 |
| GO:1990904 | 9.63e-11 |
| GO:0030529 | 9.63e-11 |
| GO:0000775 | 1.30e-10 |
| GO:0044427 | 1.34e-10 |
| GO:0005681 | 1.39e-10 |
| GO:0004386 | 2.56e-11 |
| GO:0000988 | 6.15e-11 |
| GO:0003712 | 7.06e-11 |
| GO:0003682 | 1.24e-10 |
| GO:0022890 | 1.36e-10 |
| GO:0008324 | 1.37e-10 |
| GO:0015075 | 1.38e-10 |
| GO:0000989 | 1.71e-10 |
| GO:0003723 | 1.72e-10 |
| GO:0022891 | 2.20e-10 |
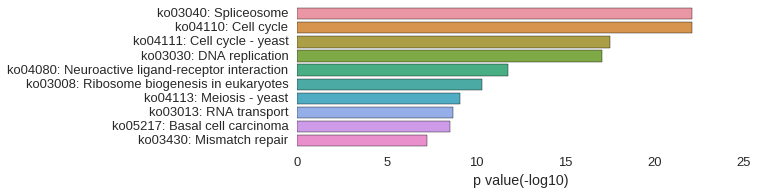
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04733
AAATGTTAAAATCTTGAAATCCTAAGATTTCCATGCACTTTTAGTATTAACGTGCACAAAGAGGCATGAT
TTATGGGATGTTTTGAGAATGAGAGGGCTGTGTAGGGGGTAGCTTGCTCAGGACCTACAGCAAAGTTCTC
TGCCTCCCCCGAAGGCTTTGAGCCCCGCTGCAGAATGGCCTCAAATGTCACGGACTGTCCCCTACCAACC
CTGCTCTCATTATAGTTTAGTCATGCTATGCTTCATTATGAACCCAAGCAGTATTCCTCATAATGAGTTT
TATTCTGTTGGCTAGTTACTTTCTGAGAACGTCCTTAGGTTTGACCAGAGTCCCCTTTTTATTTTCAGAC
GTCAACTTCATCGCAAGCCCTCCCTCCATGATTGCAGCAGGCAGTGTTGCTGCAGCGGTACAAGGACTGT
ACCTGAAAAGCACCGACAGTTGCCTCTCATCCCAGAACCTCACCAACTTCCTCTCGCAAGTCATCAGAAG
TGACCCTGTAAGTAGCGCCACCTTCCATGCATTATTCCCAGCAAGTCAATTTCTGTGTCCCTGTTACTAA
CTTTTTACTTTCTTCCACAGGACTGTCTGCGATCGTGTCAGGAACAGATCGAGTCCCTGCTGGAATCGAG
TCTCAGACAAGCTCAGCAGCACATCTCCACAGAGACCAAAAGGGTCGAGGAGGACGTGGACCTCTCTTGC
ACTCCCACAGACGTCAGAGACATTAACATTTGAAGGGATCGCTATTCATCTCGTG
AAATGTTAAAATCTTGAAATCCTAAGATTTCCATGCACTTTTAGTATTAACGTGCACAAAGAGGCATGAT
TTATGGGATGTTTTGAGAATGAGAGGGCTGTGTAGGGGGTAGCTTGCTCAGGACCTACAGCAAAGTTCTC
TGCCTCCCCCGAAGGCTTTGAGCCCCGCTGCAGAATGGCCTCAAATGTCACGGACTGTCCCCTACCAACC
CTGCTCTCATTATAGTTTAGTCATGCTATGCTTCATTATGAACCCAAGCAGTATTCCTCATAATGAGTTT
TATTCTGTTGGCTAGTTACTTTCTGAGAACGTCCTTAGGTTTGACCAGAGTCCCCTTTTTATTTTCAGAC
GTCAACTTCATCGCAAGCCCTCCCTCCATGATTGCAGCAGGCAGTGTTGCTGCAGCGGTACAAGGACTGT
ACCTGAAAAGCACCGACAGTTGCCTCTCATCCCAGAACCTCACCAACTTCCTCTCGCAAGTCATCAGAAG
TGACCCTGTAAGTAGCGCCACCTTCCATGCATTATTCCCAGCAAGTCAATTTCTGTGTCCCTGTTACTAA
CTTTTTACTTTCTTCCACAGGACTGTCTGCGATCGTGTCAGGAACAGATCGAGTCCCTGCTGGAATCGAG
TCTCAGACAAGCTCAGCAGCACATCTCCACAGAGACCAAAAGGGTCGAGGAGGACGTGGACCTCTCTTGC
ACTCCCACAGACGTCAGAGACATTAACATTTGAAGGGATCGCTATTCATCTCGTG